Sp100A promotes chromatin decondensation at a cytomegalovirus-promoter-regulated transcription site
- PMID: 23485562
- PMCID: PMC3639056
- DOI: 10.1091/mbc.E12-09-0669
Sp100A promotes chromatin decondensation at a cytomegalovirus-promoter-regulated transcription site
Abstract
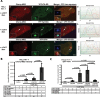
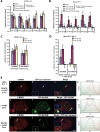
Promyelocytic leukemia nuclear bodies (PML-NBs)/nuclear domain 10s (ND10s) are nuclear structures that contain many transcriptional and chromatin regulatory factors. One of these, Sp100, is expressed from a single-copy gene and spliced into four isoforms (A, B, C, and HMG), which differentially regulate transcription. Here we evaluate Sp100 function in single cells using an inducible cytomegalovirus-promoter-regulated transgene, visualized as a chromatinized transcription site. Sp100A is the isoform most strongly recruited to the transgene array, and it significantly increases chromatin decondensation. However, Sp100A cannot overcome Daxx- and α-thalassemia mental retardation, X-linked (ATRX)-mediated transcriptional repression, which indicates that PML-NB/ND10 factors function within a regulatory hierarchy. Sp100A increases and Sp100B, which contains a SAND domain, decreases acetyl-lysine regulatory factor levels at activated sites, suggesting that Sp100 isoforms differentially regulate transcription by modulating lysine acetylation. In contrast to Daxx, ATRX, and PML, Sp100 is recruited to activated arrays in cells expressing the herpes simplex virus type 1 E3 ubiquitin ligase, ICP0, which degrades all Sp100 isoforms except unsumoylated Sp100A. The recruitment Sp100A(K297R), which cannot be sumoylated, further suggests that sumoylation plays an important role in regulating Sp100 isoform levels at transcription sites. This study provides insight into the ways in which viruses may modulate Sp100 to promote their replication cycles.
Figures
Similar articles
-
Sp100 isoform-specific regulation of human adenovirus 5 gene expression.J Virol. 2014 Jun;88(11):6076-92. doi: 10.1128/JVI.00469-14. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623443 Free PMC article.
-
Single-cell analysis of Daxx and ATRX-dependent transcriptional repression.J Cell Sci. 2012 Nov 15;125(Pt 22):5489-501. doi: 10.1242/jcs.110148. Epub 2012 Sep 12. J Cell Sci. 2012. PMID: 22976303 Free PMC article.
-
Differential functions of interferon-upregulated Sp100 isoforms: herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation.J Virol. 2009 May;83(10):5168-80. doi: 10.1128/JVI.02083-08. Epub 2009 Mar 11. J Virol. 2009. PMID: 19279115 Free PMC article.
-
The Fate of Speckled Protein 100 (Sp100) During Herpesviruses Infection.Front Cell Infect Microbiol. 2021 Feb 1;10:607526. doi: 10.3389/fcimb.2020.607526. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33598438 Free PMC article. Review.
-
Nuclear dots: actors on many stages.Immunobiology. 1997 Dec;198(1-3):307-31. doi: 10.1016/S0171-2985(97)80051-4. Immunobiology. 1997. PMID: 9442402 Review.
Cited by
-
Host cell restriction factors that limit transcription and replication of human papillomavirus.Virus Res. 2017 Mar 2;231:10-20. doi: 10.1016/j.virusres.2016.11.014. Epub 2016 Nov 15. Virus Res. 2017. PMID: 27863967 Free PMC article. Review.
-
PML Body Component Sp100A Restricts Wild-Type Herpes Simplex Virus 1 Infection.J Virol. 2022 Apr 27;96(8):e0027922. doi: 10.1128/jvi.00279-22. Epub 2022 Mar 30. J Virol. 2022. PMID: 35353002 Free PMC article.
-
RNase P protein subunit Rpp29 represses histone H3.3 nucleosome deposition.Mol Biol Cell. 2016 Apr 1;27(7):1154-69. doi: 10.1091/mbc.E15-02-0099. Epub 2016 Feb 3. Mol Biol Cell. 2016. PMID: 26842893 Free PMC article.
-
Influence of ND10 components on epigenetic determinants of early KSHV latency establishment.PLoS Pathog. 2014 Jul 17;10(7):e1004274. doi: 10.1371/journal.ppat.1004274. eCollection 2014 Jul. PLoS Pathog. 2014. PMID: 25033267 Free PMC article.
-
Viral DNA Binding Protein SUMOylation Promotes PML Nuclear Body Localization Next to Viral Replication Centers.mBio. 2020 Mar 17;11(2):e00049-20. doi: 10.1128/mBio.00049-20. mBio. 2020. PMID: 32184235 Free PMC article.
References
-
- Adler M, Tavalai N, Muller R, Stamminger T. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J Gen Virol. 2011;92:1532–1538. - PubMed
-
- Bottomley MJ, Collard MW, Huggenvik JI, Liu Z, Gibson TJ, Sattler M. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol. 2001;8:626–633. - PubMed
-
- Chelbi-Alix MK, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. - PubMed
-
- Everett RD. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. 2000;22:761–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

