H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation
- PMID: 23452851
- PMCID: PMC3588593
- DOI: 10.1016/j.cell.2013.01.052
H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation
Abstract
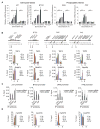
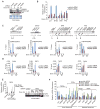
Histone modifications regulate chromatin-dependent processes, yet the mechanisms by which they contribute to specific outcomes remain unclear. H3K4me3 is a prominent histone mark that is associated with active genes and promotes transcription through interactions with effector proteins that include initiation factor TFIID. We demonstrate that H3K4me3-TAF3 interactions direct global TFIID recruitment to active genes, some of which are p53 targets. Further analyses show that (1) H3K4me3 enhances p53-dependent transcription by stimulating preinitiation complex (PIC) formation; (2) H3K4me3, through TAF3 interactions, can act either independently or cooperatively with the TATA box to direct PIC formation and transcription; and (3) H3K4me3-TAF3/TFIID interactions regulate gene-selective functions of p53 in response to genotoxic stress. Our findings indicate a mechanism by which H3K4me3 directs PIC assembly for the rapid induction of specific p53 target genes.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4.Cell. 2007 Oct 5;131(1):58-69. doi: 10.1016/j.cell.2007.08.016. Epub 2007 Sep 20. Cell. 2007. PMID: 17884155
-
Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release.Mol Cell. 2021 May 20;81(10):2166-2182.e6. doi: 10.1016/j.molcel.2021.03.008. Epub 2021 Mar 24. Mol Cell. 2021. PMID: 33765415
-
Multivalent engagement of TFIID to nucleosomes.PLoS One. 2013 Sep 11;8(9):e73495. doi: 10.1371/journal.pone.0073495. eCollection 2013. PLoS One. 2013. PMID: 24039962 Free PMC article.
-
HATs off to PIC assembly.Mol Cell. 2006 Sep 15;23(6):776-7. doi: 10.1016/j.molcel.2006.08.022. Mol Cell. 2006. PMID: 16973430 Review.
-
Broad domains of histone H3 lysine 4 trimethylation in transcriptional regulation and disease.FEBS J. 2020 Jul;287(14):2891-2902. doi: 10.1111/febs.15219. Epub 2020 Feb 4. FEBS J. 2020. PMID: 31967712 Review.
Cited by
-
Impacts of Hyperglycemia on Epigenetic Modifications in Human Gingival Fibroblasts and Gingiva in Diabetic Rats.Int J Mol Sci. 2024 Oct 12;25(20):10979. doi: 10.3390/ijms252010979. Int J Mol Sci. 2024. PMID: 39456763 Free PMC article.
-
The evolving metabolic landscape of chromatin biology and epigenetics.Nat Rev Genet. 2020 Dec;21(12):737-753. doi: 10.1038/s41576-020-0270-8. Epub 2020 Sep 9. Nat Rev Genet. 2020. PMID: 32908249 Free PMC article. Review.
-
RNA interacts with topoisomerase I to adjust DNA topology.Mol Cell. 2024 Sep 5;84(17):3192-3208.e11. doi: 10.1016/j.molcel.2024.07.032. Epub 2024 Aug 21. Mol Cell. 2024. PMID: 39173639
-
A Genome-Wide Chronological Study of Gene Expression and Two Histone Modifications, H3K4me3 and H3K9ac, during Developmental Leaf Senescence.Plant Physiol. 2015 Aug;168(4):1246-61. doi: 10.1104/pp.114.252999. Epub 2015 Mar 23. Plant Physiol. 2015. PMID: 25802367 Free PMC article.
-
Rebooting the Epigenomes during Mammalian Early Embryogenesis.Stem Cell Reports. 2020 Dec 8;15(6):1158-1175. doi: 10.1016/j.stemcr.2020.09.005. Epub 2020 Oct 8. Stem Cell Reports. 2020. PMID: 33035464 Free PMC article. Review.
References
-
- An W, Roeder RG. Reconstitution and transcriptional analysis of chromatin in vitro. Methods Enzymol. 2004;377:460–474. - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
-
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. - PubMed
-
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

