DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes
- PMID: 23422670
- PMCID: PMC3633418
- DOI: 10.1038/ncomms2502
DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes
Abstract
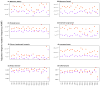
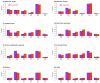
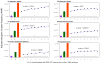
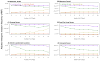
Single-nucleotide substitutions are a defining characteristic of cancer genomes. Many single-nucleotide substitutions in cancer genomes arise because of errors in DNA replication, which is spatio-temporally stratified. Here we propose that DNA replication patterns help shape the mutational landscapes of normal and cancer genomes. Using data on five fully sequenced cancer types and two personal genomes, we determined that the frequency of intergenic single-nucleotide substitution is significantly higher in late DNA replication timing regions, even after controlling for a number of genomic features. Furthermore, some substitution signatures are more frequent in certain DNA replication timing zones. Finally, integrating data on higher-order nuclear organization, we found that genomic regions in close spatial proximity to late-replicating domains display similar mutation spectra as the late-replicating regions themselves. These data suggest that DNA replication timing together with higher-order genomic organization contribute to the patterns of single-nucleotide substitution in normal and cancer genomes.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes.Nat Biotechnol. 2011 Nov 20;29(12):1103-8. doi: 10.1038/nbt.2030. Nat Biotechnol. 2011. PMID: 22101487 Free PMC article.
-
DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes.Nat Commun. 2012;3:1004. doi: 10.1038/ncomms1982. Nat Commun. 2012. PMID: 22893128
-
Nuclear topology modulates the mutational landscapes of cancer genomes.Nat Struct Mol Biol. 2017 Nov;24(11):1000-1006. doi: 10.1038/nsmb.3474. Epub 2017 Oct 2. Nat Struct Mol Biol. 2017. PMID: 28967881 Free PMC article.
-
DNA replication timing, genome stability and cancer: late and/or delayed DNA replication timing is associated with increased genomic instability.Semin Cancer Biol. 2013 Apr;23(2):80-9. doi: 10.1016/j.semcancer.2013.01.001. Epub 2013 Jan 14. Semin Cancer Biol. 2013. PMID: 23327985 Free PMC article. Review.
-
Replication timing and nuclear structure.Curr Opin Cell Biol. 2018 Jun;52:43-50. doi: 10.1016/j.ceb.2018.01.004. Epub 2018 Feb 4. Curr Opin Cell Biol. 2018. PMID: 29414592 Free PMC article. Review.
Cited by
-
A computational strategy to adjust for copy number in tumor Hi-C data.Bioinformatics. 2016 Dec 15;32(24):3695-3701. doi: 10.1093/bioinformatics/btw540. Epub 2016 Aug 16. Bioinformatics. 2016. PMID: 27531101 Free PMC article.
-
Differential DNA mismatch repair underlies mutation rate variation across the human genome.Nature. 2015 May 7;521(7550):81-4. doi: 10.1038/nature14173. Epub 2015 Feb 23. Nature. 2015. PMID: 25707793 Free PMC article.
-
Spatially coordinated replication and minimization of expression noise constrain three-dimensional organization of yeast genome.DNA Res. 2016 Apr;23(2):155-69. doi: 10.1093/dnares/dsw005. Epub 2016 Feb 29. DNA Res. 2016. PMID: 26932984 Free PMC article.
-
Loss of heterozygosity preferentially occurs in early replicating regions in cancer genomes.Nucleic Acids Res. 2013 Sep;41(16):7615-24. doi: 10.1093/nar/gkt552. Epub 2013 Jun 22. Nucleic Acids Res. 2013. PMID: 23793816 Free PMC article.
-
Coordination of DNA damage tolerance mechanisms with cell cycle progression in fission yeast.Cell Cycle. 2016;15(2):261-73. doi: 10.1080/15384101.2015.1121353. Cell Cycle. 2016. PMID: 26652183 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

