PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress
- PMID: 23416000
- PMCID: PMC3708305
- DOI: 10.1016/j.ccr.2012.11.020
PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress
Abstract
Cancer cells reprogram their metabolism using different strategies to meet energy and anabolic demands to maintain growth and survival. Understanding the molecular and genetic determinants of these metabolic programs is critical to successfully exploit them for therapy. Here, we report that the oncogenic melanocyte lineage-specification transcription factor MITF drives PGC1α (PPARGC1A) overexpression in a subset of human melanomas and derived cell lines. Functionally, PGC1α positive melanoma cells exhibit increased mitochondrial energy metabolism and reactive oxygen species (ROS) detoxification capacities that enable survival under oxidative stress conditions. Conversely, PGC1α negative melanoma cells are more glycolytic and sensitive to ROS-inducing drugs. These results demonstrate that differences in PGC1α levels in melanoma tumors have a profound impact in their metabolism, biology, and drug sensitivity.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
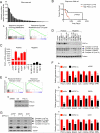
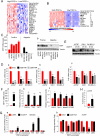
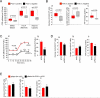


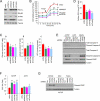


Comment in
-
Melanoma: Horses for courses.Nat Rev Cancer. 2013 Apr;13(4):222. doi: 10.1038/nrc3491. Epub 2013 Feb 28. Nat Rev Cancer. 2013. PMID: 23446548 No abstract available.
-
Targeting oxidative phosphorylation: why, when, and how.Cancer Cell. 2013 Mar 18;23(3):263-4. doi: 10.1016/j.ccr.2013.02.015. Cancer Cell. 2013. PMID: 23518341
Similar articles
-
Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF.Cancer Cell. 2013 Mar 18;23(3):302-15. doi: 10.1016/j.ccr.2013.02.003. Epub 2013 Mar 7. Cancer Cell. 2013. PMID: 23477830 Free PMC article.
-
Targeting mitochondrial oxidative metabolism in melanoma causes metabolic compensation through glucose and glutamine utilization.Cancer Res. 2014 Jul 1;74(13):3535-45. doi: 10.1158/0008-5472.CAN-13-2893-T. Epub 2014 May 8. Cancer Res. 2014. PMID: 24812272
-
PGC-1 coactivators regulate MITF and the tanning response.Mol Cell. 2013 Jan 10;49(1):145-57. doi: 10.1016/j.molcel.2012.10.027. Epub 2012 Nov 29. Mol Cell. 2013. PMID: 23201126 Free PMC article.
-
Mitochondrial function in melanoma.Arch Biochem Biophys. 2014 Dec 1;563:56-9. doi: 10.1016/j.abb.2014.06.028. Epub 2014 Jul 2. Arch Biochem Biophys. 2014. PMID: 24997363 Review.
-
PGC1α and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders.J Cell Sci. 2012 Nov 1;125(Pt 21):4963-71. doi: 10.1242/jcs.113662. J Cell Sci. 2012. PMID: 23277535 Review.
Cited by
-
Hypoxia Induces Mitochondrial Defect That Promotes T Cell Exhaustion in Tumor Microenvironment Through MYC-Regulated Pathways.Front Immunol. 2020 Aug 21;11:1906. doi: 10.3389/fimmu.2020.01906. eCollection 2020. Front Immunol. 2020. PMID: 32973789 Free PMC article.
-
Omics analyses of a somatic Trp53R245W/+ breast cancer model identify cooperating driver events activating PI3K/AKT/mTOR signaling.Proc Natl Acad Sci U S A. 2022 Nov 8;119(45):e2210618119. doi: 10.1073/pnas.2210618119. Epub 2022 Nov 2. Proc Natl Acad Sci U S A. 2022. PMID: 36322759 Free PMC article.
-
MCU controls melanoma progression through a redox-controlled phenotype switch.EMBO Rep. 2022 Nov 7;23(11):e54746. doi: 10.15252/embr.202254746. Epub 2022 Sep 26. EMBO Rep. 2022. PMID: 36156348 Free PMC article.
-
Coupling of LETM1 up-regulation with oxidative phosphorylation and platelet-derived growth factor receptor signaling via YAP1 transactivation.Oncotarget. 2016 Oct 11;7(41):66728-66739. doi: 10.18632/oncotarget.11456. Oncotarget. 2016. PMID: 27556512 Free PMC article.
-
BRAF - a tumour-agnostic drug target with lineage-specific dependencies.Nat Rev Clin Oncol. 2024 Mar;21(3):224-247. doi: 10.1038/s41571-023-00852-0. Epub 2024 Jan 26. Nat Rev Clin Oncol. 2024. PMID: 38278874 Review.
References
-
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d'Hayer B, Mohamdi H, Remenieras A, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. - PubMed
-
- Bogunovic D, O'Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, Darvishian F, Berman R, Shapiro R, Pavlick AC, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20429–20434. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

