Searching the coding region for microRNA targets
- PMID: 23404894
- PMCID: PMC3677256
- DOI: 10.1261/rna.035634.112
Searching the coding region for microRNA targets
Abstract
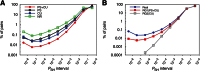
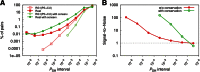
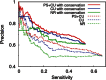
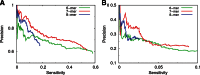
Finding microRNA targets in the coding region is difficult due to the overwhelming signal encoding the amino acid sequence. Here, we introduce an algorithm (called PACCMIT-CDS) that finds potential microRNA targets within coding sequences by searching for conserved motifs that are complementary to the microRNA seed region and also overrepresented in comparison with a background model preserving both codon usage and amino acid sequence. Precision and sensitivity of PACCMIT-CDS are evaluated using PAR-CLIP and proteomics data sets. Thanks to the properly constructed background, the new algorithm achieves a lower rate of false positives and better ranking of predictions than do currently available algorithms, which were designed to find microRNA targets within 3' UTRs.
Figures
Similar articles
-
PACCMIT/PACCMIT-CDS: identifying microRNA targets in 3' UTRs and coding sequences.Nucleic Acids Res. 2015 Jul 1;43(W1):W474-9. doi: 10.1093/nar/gkv457. Epub 2015 May 6. Nucleic Acids Res. 2015. PMID: 25948580 Free PMC article.
-
Optimal use of conservation and accessibility filters in microRNA target prediction.PLoS One. 2012;7(2):e32208. doi: 10.1371/journal.pone.0032208. Epub 2012 Feb 27. PLoS One. 2012. PMID: 22384176 Free PMC article.
-
Functional microRNA targets in protein coding sequences.Bioinformatics. 2012 Mar 15;28(6):771-6. doi: 10.1093/bioinformatics/bts043. Epub 2012 Jan 27. Bioinformatics. 2012. PMID: 22285563
-
MicroRNA target finding by comparative genomics.Methods Mol Biol. 2014;1097:457-76. doi: 10.1007/978-1-62703-709-9_21. Methods Mol Biol. 2014. PMID: 24639172 Review.
-
Experimental identification of microRNA targets.Gene. 2010 Feb 1;451(1-2):1-5. doi: 10.1016/j.gene.2009.11.008. Epub 2009 Nov 24. Gene. 2010. PMID: 19944134 Review.
Cited by
-
Synonymous Variants: Necessary Nuance in Our Understanding of Cancer Drivers and Treatment Outcomes.J Natl Cancer Inst. 2022 Aug 8;114(8):1072-1094. doi: 10.1093/jnci/djac090. J Natl Cancer Inst. 2022. PMID: 35477782 Free PMC article. Review.
-
Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development.Nat Neurosci. 2018 Dec;21(12):1784-1792. doi: 10.1038/s41593-018-0265-3. Epub 2018 Nov 19. Nat Neurosci. 2018. PMID: 30455455 Free PMC article.
-
Dietary plant miRNAs as an augmented therapy: cross-kingdom gene regulation.RNA Biol. 2018;15(12):1433-1439. doi: 10.1080/15476286.2018.1551693. Epub 2018 Dec 10. RNA Biol. 2018. PMID: 30474479 Free PMC article. Review.
-
In silico methods for predicting functional synonymous variants.Genome Biol. 2023 May 22;24(1):126. doi: 10.1186/s13059-023-02966-1. Genome Biol. 2023. PMID: 37217943 Free PMC article. Review.
-
Identification and Characterization of MicroRNAs Involving in Initial Sex Differentiation of Chlamys farreri Gonads.Biology (Basel). 2022 Mar 16;11(3):456. doi: 10.3390/biology11030456. Biology (Basel). 2022. PMID: 35336829 Free PMC article.
References
-
- Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG 2009. Lost in translation: An assessment and perspective for computational microRNA target identification. Bioinformatics 25: 3049–3055 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
