Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study
- PMID: 23382473
- PMCID: PMC3595425
- DOI: 10.1200/JCO.2012.44.3184
Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study
Abstract
Purpose: To identify a robust prognostic gene expression signature as an independent predictor of survival of patients with acute myeloid leukemia (AML) and use it to improve established risk classification.
Patients and methods: Four independent sets totaling 499 patients with AML carrying various cytogenetic and molecular abnormalities were used as training sets. Two independent patient sets composed of 825 patients were used as validation sets. Notably, patients from different sets were treated with different protocols, and their gene expression profiles were derived using different microarray platforms. Cox regression and Kaplan-Meier methods were used for survival analyses.
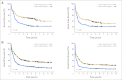
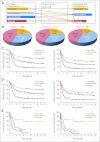
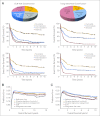
Results: A prognostic signature composed of 24 genes was derived from a meta-analysis of Cox regression values of each gene across the four training sets. In multivariable models, a higher sum value of the 24-gene signature was an independent predictor of shorter overall (OS) and event-free survival (EFS) in both training and validation sets (P < .01). Moreover, this signature could substantially improve the European LeukemiaNet (ELN) risk classification of AML, and patients in three new risk groups classified by the integrated risk classification showed significantly (P < .001) distinct OS and EFS.
Conclusion: Despite different treatment protocols applied to patients and use of different microarray platforms for expression profiling, a common prognostic gene signature was identified as an independent predictor of survival of patients with AML. The integrated risk classification incorporating this gene signature provides a better framework for risk stratification and outcome prediction than the ELN classification.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures

Similar articles
-
TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study.J Clin Oncol. 2011 Apr 1;29(10):1373-81. doi: 10.1200/JCO.2010.32.7742. Epub 2011 Feb 22. J Clin Oncol. 2011. PMID: 21343549 Free PMC article. Clinical Trial.
-
Incorporation of long non-coding RNA expression profile in the 2017 ELN risk classification can improve prognostic prediction of acute myeloid leukemia patients.EBioMedicine. 2019 Feb;40:240-250. doi: 10.1016/j.ebiom.2019.01.022. Epub 2019 Jan 17. EBioMedicine. 2019. PMID: 30662003 Free PMC article.
-
Development and validation of a promising 5-gene prognostic model for pediatric acute myeloid leukemia.Mol Biomed. 2024 Jan 2;5(1):1. doi: 10.1186/s43556-023-00162-y. Mol Biomed. 2024. PMID: 38163849 Free PMC article.
-
Stratification of acute myeloid leukemia based on gene expression profiles.Int J Hematol. 2004 Dec;80(5):389-94. doi: 10.1532/ijh97.04111. Int J Hematol. 2004. PMID: 15646648 Review.
-
A Focus on Intermediate-Risk Acute Myeloid Leukemia: Sub-Classification Updates and Therapeutic Challenges.Cancers (Basel). 2022 Aug 28;14(17):4166. doi: 10.3390/cancers14174166. Cancers (Basel). 2022. PMID: 36077703 Free PMC article. Review.
Cited by
-
Transcriptionally imprinted glycomic signatures of acute myeloid leukemia.Cell Biosci. 2023 Feb 14;13(1):31. doi: 10.1186/s13578-023-00981-0. Cell Biosci. 2023. PMID: 36788594 Free PMC article.
-
Pan-cancer analysis identifies CD300 molecules as potential immune regulators and promising therapeutic targets in acute myeloid leukemia.Cancer Med. 2023 Jan;12(1):789-807. doi: 10.1002/cam4.4905. Epub 2022 May 31. Cancer Med. 2023. PMID: 35642341 Free PMC article.
-
Pan-cancer analysis revealing that PTPN2 is an indicator of risk stratification for acute myeloid leukemia.Sci Rep. 2023 Oct 26;13(1):18372. doi: 10.1038/s41598-023-44892-z. Sci Rep. 2023. PMID: 37884566 Free PMC article.
-
Allogeneic stem cell transplantation for acute myeloid leukemia in first complete remission: are we closer to knowing who needs it?Curr Hematol Malig Rep. 2014 Jun;9(2):128-37. doi: 10.1007/s11899-014-0207-4. Curr Hematol Malig Rep. 2014. PMID: 24664820 Review.
-
CEBPE expression is an independent prognostic factor for acute myeloid leukemia.J Transl Med. 2019 Jun 4;17(1):188. doi: 10.1186/s12967-019-1944-x. J Transl Med. 2019. PMID: 31164135 Free PMC article.
References
-
- Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. - PubMed
-
- Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. - PubMed
-
- Marcucci G, Mrózek K, Bloomfield CD. Molecular heterogeneity and prognostic biomarkers in adults with acute myeloid leukemia and normal cytogenetics. Curr Opin Hematol. 2005;12:68–75. - PubMed
-
- Grimwade D, Mrózek K. Diagnostic and prognostic value of cytogenetics in acute myeloid leukemia. Hematol Oncol Clin North Am. 2011;25:1135–1161. - PubMed
-
- Smith ML, Hills RK, Grimwade D. Independent prognostic variables in acute myeloid leukaemia. Blood Rev. 2011;25:39–51. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

