Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]
- PMID: 23382459
- PMCID: PMC3628449
- DOI: 10.1093/dnares/dst002
Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]
Abstract
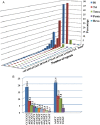
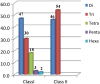
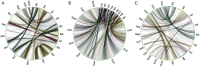
The availability of well-validated informative co-dominant microsatellite markers and saturated genetic linkage map has been limited in foxtail millet (Setaria italica L.). In view of this, we conducted a genome-wide analysis and identified 28 342 microsatellite repeat-motifs spanning 405.3 Mb of foxtail millet genome. The trinucleotide repeats (∼48%) was prevalent when compared with dinucleotide repeats (∼46%). Of the 28 342 microsatellites, 21 294 (∼75%) primer pairs were successfully designed, and a total of 15 573 markers were physically mapped on 9 chromosomes of foxtail millet. About 159 markers were validated successfully in 8 accessions of Setaria sp. with ∼67% polymorphic potential. The high percentage (89.3%) of cross-genera transferability across millet and non-millet species with higher transferability percentage in bioenergy grasses (∼79%, Switchgrass and ∼93%, Pearl millet) signifies their importance in studying the bioenergy grasses. In silico comparative mapping of 15 573 foxtail millet microsatellite markers against the mapping data of sorghum (16.9%), maize (14.5%) and rice (6.4%) indicated syntenic relationships among the chromosomes of foxtail millet and target species. The results, thus, demonstrate the immense applicability of developed microsatellite markers in germplasm characterization, phylogenetics, construction of genetic linkage map for gene/quantitative trait loci discovery, comparative mapping in foxtail millet, including other millets and bioenergy grass species.
Figures

Similar articles
-
Development of eSSR-Markers in Setaria italica and Their Applicability in Studying Genetic Diversity, Cross-Transferability and Comparative Mapping in Millet and Non-Millet Species.PLoS One. 2013 Jun 21;8(6):e67742. doi: 10.1371/journal.pone.0067742. Print 2013. PLoS One. 2013. PMID: 23805325 Free PMC article.
-
Sequence-based novel genomic microsatellite markers for robust genotyping purposes in foxtail millet [Setaria italica (L.) P. Beauv].Plant Cell Rep. 2012 Feb;31(2):323-37. doi: 10.1007/s00299-011-1168-x. Epub 2011 Oct 13. Plant Cell Rep. 2012. PMID: 21993813
-
Development of 5123 intron-length polymorphic markers for large-scale genotyping applications in foxtail millet.DNA Res. 2014 Feb;21(1):41-52. doi: 10.1093/dnares/dst039. Epub 2013 Oct 1. DNA Res. 2014. PMID: 24086082 Free PMC article.
-
Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses.Theor Appl Genet. 2015 Jan;128(1):1-14. doi: 10.1007/s00122-014-2399-3. Epub 2014 Sep 20. Theor Appl Genet. 2015. PMID: 25239219 Review.
-
Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses.Crit Rev Biotechnol. 2013 Sep;33(3):328-43. doi: 10.3109/07388551.2012.716809. Epub 2012 Sep 18. Crit Rev Biotechnol. 2013. PMID: 22985089 Review.
Cited by
-
Multi-omics intervention in Setaria to dissect climate-resilient traits: Progress and prospects.Front Plant Sci. 2022 Aug 31;13:892736. doi: 10.3389/fpls.2022.892736. eCollection 2022. Front Plant Sci. 2022. PMID: 36119586 Free PMC article. Review.
-
Genome-wide distribution comparative and composition analysis of the SSRs in Poaceae.BMC Genet. 2015 Feb 15;16:18. doi: 10.1186/s12863-015-0178-z. BMC Genet. 2015. PMID: 25886726 Free PMC article.
-
Population structure and association mapping of yield contributing agronomic traits in foxtail millet.Plant Cell Rep. 2014 Jun;33(6):881-93. doi: 10.1007/s00299-014-1564-0. Epub 2014 Jan 12. Plant Cell Rep. 2014. PMID: 24413764
-
FmTFDb: a foxtail millet transcription factors database for expediting functional genomics in millets.Mol Biol Rep. 2014 Oct;41(10):6343-8. doi: 10.1007/s11033-014-3574-y. Epub 2014 Jul 9. Mol Biol Rep. 2014. PMID: 25005261
-
Molecular Approaches to Understand Nutritional Potential of Coarse Cereals.Curr Genomics. 2016 Jun;17(3):177-92. doi: 10.2174/1389202917666160202215308. Curr Genomics. 2016. PMID: 27252585 Free PMC article.
References
-
- Lata C., Gupta S., Prasad M. Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit. Rev. Biotechnol. 2012 DOI:10.3109/07388551.2012.716809. (September 18, 2012) - DOI - PubMed
-
- Li P., Brutnell T.P. Setaria viridis and Setaria italica, model genetic systems for Panicoid grasses. J. Exp. Bot. 2011;62:3031–37. - PubMed
-
- Bennetzen J.L., Wang J.S.H., Percifield R., et al. Reference genome sequence of the model plant Setaria. Nature Biotech. 2012;30:555–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

