Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response
- PMID: 23382245
- PMCID: PMC3581903
- DOI: 10.1073/pnas.1213737110
Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response
Abstract
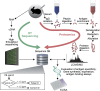
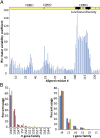
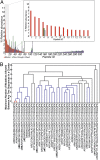
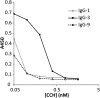
We have developed and validated a methodology for determining the antibody composition of the polyclonal serum response after immunization. Pepsin-digested serum IgGs were subjected to standard antigen-affinity chromatography, and resulting elution, wash, and flow-through fractions were analyzed by bottom-up, liquid chromatography-high-resolution tandem mass spectrometry. Identification of individual monoclonal antibodies required the generation of a database of IgG variable gene (V-gene) sequences constructed by NextGen sequencing of mature B cells. Antibody V-gene sequences are characterized by short complementarity determining regions (CDRs) of high diversity adjacent to framework regions shared across thousands of IgGs, greatly complicating the identification of antigen-specific IgGs from proteomically observed peptides. By mapping peptides marking unique V(H) CDRH3 sequences, we identified a set of V-genes heavily enriched in the affinity chromatography elution, constituting the serum polyclonal response. After booster immunization in a rabbit, we find that the antigen-specific serum immune response is oligoclonal, comprising antibodies encoding 34 different CDRH3s that group into 30 distinct antibody V(H) clonotypes. Of these 34 CDRH3s, 12 account for ∼60% of the antigen-specific CDRH3 peptide mass spectral counts. For comparison, antibodies with 18 different CDRH3s (12 clonotypes) were represented in the antigen-specific IgG fraction from an unimmunized rabbit that fortuitously displayed a moderate titer for BSA. Proteomically identified antibodies were synthesized and shown to display subnanomolar affinities. The ability to deconvolute the polyclonal serum response is likely to be of key importance for analyzing antibody responses after vaccination and for more completely understanding adaptive immune responses in health and disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Trimeric gp120-specific bovine monoclonal antibodies require cysteine and aromatic residues in CDRH3 for high affinity binding to HIV Env.MAbs. 2017 Apr;9(3):550-566. doi: 10.1080/19420862.2016.1270491. Epub 2016 Dec 20. MAbs. 2017. PMID: 27996375 Free PMC article.
-
A proteomics approach for the identification and cloning of monoclonal antibodies from serum.Nat Biotechnol. 2012 Mar 25;30(5):447-52. doi: 10.1038/nbt.2167. Nat Biotechnol. 2012. PMID: 22446692
-
Proteomic identification of monoclonal antibodies from serum.Anal Chem. 2014 May 20;86(10):4758-66. doi: 10.1021/ac4037679. Epub 2014 May 1. Anal Chem. 2014. PMID: 24684310 Free PMC article.
-
SDR grafting--a new approach to antibody humanization.Methods. 2005 May;36(1):25-34. doi: 10.1016/j.ymeth.2005.01.003. Methods. 2005. PMID: 15848072 Review.
-
Length distribution of CDRH3 in antibodies.Proteins. 1993 May;16(1):1-7. doi: 10.1002/prot.340160102. Proteins. 1993. PMID: 8497480 Review.
Cited by
-
Immunoglobulin Classification Using the Colored Antibody Graph.J Comput Biol. 2016 Jun;23(6):483-94. doi: 10.1089/cmb.2016.0010. Epub 2016 May 5. J Comput Biol. 2016. PMID: 27149636 Free PMC article.
-
A quantitative tool to distinguish isobaric leucine and isoleucine residues for mass spectrometry-based de novo monoclonal antibody sequencing.J Am Soc Mass Spectrom. 2014 Jul;25(7):1228-36. doi: 10.1007/s13361-014-0892-1. Epub 2014 May 21. J Am Soc Mass Spectrom. 2014. PMID: 24845350
-
Coupling enrichment methods with proteomics for understanding and treating disease.Proteomics Clin Appl. 2015 Feb;9(1-2):33-47. doi: 10.1002/prca.201400097. Epub 2015 Jan 19. Proteomics Clin Appl. 2015. PMID: 25523641 Free PMC article. Review.
-
The evolution within us.Philos Trans R Soc Lond B Biol Sci. 2015 Sep 5;370(1676):20140235. doi: 10.1098/rstb.2014.0235. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26194749 Free PMC article. Review.
-
Quantitative Mass Spectrometric Analysis of Autoantibodies as a Paradigm Shift in Autoimmune Serology.Front Immunol. 2019 Dec 4;10:2845. doi: 10.3389/fimmu.2019.02845. eCollection 2019. Front Immunol. 2019. PMID: 31867009 Free PMC article. No abstract available.
References
-
- Browning CH. Emil Behring and Paul Ehrlich: Their contributions to science. Nature. 1955;175(4457):570–575. - PubMed
-
- Kantha SS. A centennial review; the 1890 tetanus antitoxin paper of von Behring and Kitasato and the related developments. Keio J Med. 1991;40(1):35–39. - PubMed
-
- Radbruch A, et al. Competence and competition: The challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6(10):741–750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

