Transforming mutations of RAC guanosine triphosphatases in human cancers
- PMID: 23382236
- PMCID: PMC3581941
- DOI: 10.1073/pnas.1216141110
Transforming mutations of RAC guanosine triphosphatases in human cancers
Abstract
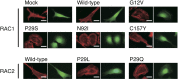
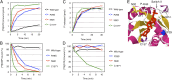
Members of the RAS superfamily of small guanosine triphosphatases (GTPases) transition between GDP-bound, inactive and GTP-bound, active states and thereby function as binary switches in the regulation of various cellular activities. Whereas HRAS, NRAS, and KRAS frequently acquire transforming missense mutations in human cancer, little is known of the oncogenic roles of other small GTPases, including Ras-related C3 botulinum toxin substrate (RAC) proteins. We show that the human sarcoma cell line HT1080 harbors both NRAS(Q61K) and RAC1(N92I) mutant proteins. Whereas both of these mutants were able to transform fibroblasts, knockdown experiments indicated that RAC1(N92I) may be the essential growth driver for this cell line. Screening for RAC1, RAC2, or RAC3 mutations in cell lines and public databases identified several missense mutations for RAC1 and RAC2, with some of the mutant proteins, including RAC1(P29S), RAC1(C157Y), RAC2(P29L), and RAC2(P29Q), being found to be activated and transforming. P29S, N92I, and C157Y mutants of RAC1 were shown to exist preferentially in the GTP-bound state as a result of a rapid transition from the GDP-bound state, rather than as a result of a reduced intrinsic GTPase activity. Activating mutations of RAC GTPases were thus found in a wide variety of human cancers at a low frequency; however, given their marked transforming ability, the mutant proteins are potential targets for the development of new therapeutic agents.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Structural requirements for PAK activation by Rac GTPases.J Biol Chem. 1998 Aug 21;273(34):21512-8. doi: 10.1074/jbc.273.34.21512. J Biol Chem. 1998. PMID: 9705280
-
Assessing the mechanism of fast-cycling cancer-associated mutations of Rac1 small Rho GTPase.Protein Sci. 2024 Apr;33(4):e4939. doi: 10.1002/pro.4939. Protein Sci. 2024. PMID: 38501467
-
Purification and biochemical properties of Rac1, 2, 3 and the splice variant Rac1b.Methods Enzymol. 2006;406:1-11. doi: 10.1016/S0076-6879(06)06001-0. Methods Enzymol. 2006. PMID: 16472645
-
Rac GTPases in human diseases.Dis Markers. 2010;29(3-4):177-87. doi: 10.3233/DMA-2010-0738. Dis Markers. 2010. PMID: 21178276 Free PMC article. Review.
-
Hematopoietic-specific Rho GTPases Rac2 and RhoH and human blood disorders.Exp Cell Res. 2013 Sep 10;319(15):2375-83. doi: 10.1016/j.yexcr.2013.07.002. Epub 2013 Jul 11. Exp Cell Res. 2013. PMID: 23850828 Free PMC article. Review.
Cited by
-
Di-Ras2 Protein Forms a Complex with SmgGDS Protein in Brain Cytosol in Order to Be in a Low Affinity State for Guanine Nucleotides.J Biol Chem. 2015 Aug 14;290(33):20245-56. doi: 10.1074/jbc.M115.637769. Epub 2015 Jul 6. J Biol Chem. 2015. PMID: 26149690 Free PMC article.
-
Wavelet coherence phase analysis decodes the universal switching mechanism of Ras GTPase superfamily.iScience. 2023 Jun 7;26(7):107031. doi: 10.1016/j.isci.2023.107031. eCollection 2023 Jul 21. iScience. 2023. PMID: 37448564 Free PMC article.
-
Understanding cancer drug resistance with Sleeping Beauty functional genomic screens: Application to MAPK inhibition in cutaneous melanoma.iScience. 2023 Aug 31;26(10):107805. doi: 10.1016/j.isci.2023.107805. eCollection 2023 Oct 20. iScience. 2023. PMID: 37860756 Free PMC article.
-
Rho GTPases modulate malignant transformation of tumor cells.Small GTPases. 2014;5:e29019. doi: 10.4161/sgtp.29019. Epub 2014 May 8. Small GTPases. 2014. PMID: 25036871 Free PMC article. Review.
-
Insights into the biological functions of Dock family guanine nucleotide exchange factors.Genes Dev. 2014 Mar 15;28(6):533-47. doi: 10.1101/gad.236349.113. Genes Dev. 2014. PMID: 24637113 Free PMC article.
References
-
- Druker BJ, et al. IRIS Investigators Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. - PubMed
-
- Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

