A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α
- PMID: 23381944
- PMCID: PMC3598078
- DOI: 10.1002/emmm.201201161
A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α
Abstract
Emerging evidence points to aberrant regulation of translation as a driver of cell transformation in cancer. Given the direct control of translation by tRNA modifications, tRNA modifying enzymes may function as regulators of cancer progression. Here, we show that a tRNA methyltransferase 9-like (hTRM9L/KIAA1456) mRNA is down-regulated in breast, bladder, colorectal, cervix and testicular carcinomas. In the aggressive SW620 and HCT116 colon carcinoma cell lines, hTRM9L is silenced and its re-expression and methyltransferase activity dramatically suppressed tumour growth in vivo. This growth inhibition was linked to decreased proliferation, senescence-like G0/G1-arrest and up-regulation of the RB interacting protein LIN9. Additionally, SW620 cells re-expressing hTRM9L did not respond to hypoxia via HIF1-α-dependent induction of GLUT1. Importantly, hTRM9L-negative tumours were highly sensitive to aminoglycoside antibiotics and this was associated with altered tRNA modification levels compared to antibiotic resistant hTRM9L-expressing SW620 cells. Our study links hTRM9L and tRNA modifications to inhibition of tumour growth via LIN9 and HIF1-α-dependent mechanisms. It also suggests that aminoglycoside antibiotics may be useful to treat hTRM9L-deficient tumours.
Copyright © 2013 The Authors. Published by John Wiley and Sons, Ltd on behalf of EMBO.
Figures

A. Annotated gene tree of yeast Trm9 homologs. Two yTrm9 homologs are found in humans, KIAA1456(hTRM9L) and hALKBH8. In addition to the SAM-dependent Methyltransferase domain, hALKBH8 contains a RNA Recognition Motif and an AlkB domain.
B. KIAA1456 (hTRM9L) expression profiling of Origene Tissue Scan qPCR array covering 18 different cancer types was used to identify significant differences in expression of hTRM9L in these cancer tissues compared to normal tissues. Significant differences in hTRM9L transcripts were evaluated and scored (Log10 p-value).
C. hTRM9L expression was analysed in various colorectal cancer cell lines. RNA was isolated from SW620, SW480, HT29, HCT116 and SW1116 cell lines and hTRM9L transcript levels were quantitated (ΔΔCT method) by qPCR.
D,E. HCT116 or SW620 cells were treated with 10 µM 5-aza-dC over a period of 7 days with the culture being replaced every 24 h with fresh containing 5-aza-dC. RNA was isolated and hTRM9L expression was determined by qPCR analysis.
F. Mock or 5-aza-dC treated SW620 cells (5 × 105) were inoculated on CAM and grown for 7 days in vivo. Tumours were excised, minced and collagenased and the number of cells per tumour nodule was counted. Statistical significance determined by paired student's t-test.
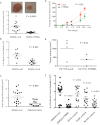
A. 5 × 105 SW620 cells expressing either LacZ or hTRM9L were inoculated on CAM and tumours were analysed after 7 days in vivo. Tumours were excised, minced and collagenased and the number of cells per tumour nodule was counted.
B,C. Experiments similar to A were performed with (B) yTrm9 and (C) mouse Alkbh8.
D,E. SW620 cells expressing either LacZ or hTRM9L were inoculated in nude mice and tumours were measured every 2 days, up to 11 days. Average data for all days is plotted in (D) and day 11 data for all tumours is shown in (E). HCT116 cells expressing either LacZ or hTRM9L were inoculated in nude mice and tumours were measured every 2 days, with 12-day data shown.
F. SW620 cells expressing either LacZ, hTRM9L or its single amino acid mutants hTRM9L-D91R, hTRM9L-I108N and hTRM9L-R410E were inoculated on CAM and tumours were analysed after 7 days. Tumours were excised, minced and collagenased and the number of cells per tumour nodule was counted.

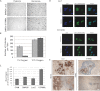
A–E. Tumour nodules of SW620 parent, LacZ and hTRM9L expressing cells were harvested 7 days post-inoculation, fixed and prepared for (A) cleaved caspase 3, (B) LC-3 (C) phospho-Histone3 (pH3), (D) p21 and (E) p27 immunohistochemistry.
F. SW620-GFP and hTRM9L expressing cells from tumour nodules were harvested after several time points as indicated and stained for acidic β-gal. Statistical significance determined by paired student's t-test.
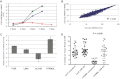
SW620 (blue circle), LacZ (green square) and hTRM9L (red triangle) tumours were harvested after several time points as indicated, minced, collagenased and quantitated as before.
The intensity values measured by microarray analysis are represented in a scatter plot comparing expression patterns of SW620-LacZ (x-axis) and SW620-hTRM9L (y-axis) of 3-day-old tumours grown on the CAM. Each gene in the microarray is represented by a dot with coordinates consisting of average gene expression measured from three different replicates of RNA samples. A linear regression line is overlaid with the scatter plot and the regression equation is displayed. Outliers represent up-regulated or down-regulated genes.
Real-time PCR validation of POR, LIN9, ALAS1, and hTRM9L respective expression levels. RNA was isolated from a GFP-positive population isolated from 3 day tumours grown on the CAM and amplified using MessageBOOSTER™ cDNA Synthesis Kit. Relative quantification is presented after normalization with GAPDH.
Cells with a knock down of LIN9 or a scrambled control, in hTRM9L and LacZ expressing variants, were assayed for their tumour forming capacity, along with the parental cells. Statistical significance determined by paired student's t-test.
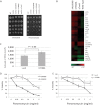
A,B. SW620-LacZ (light grey bars) and SW620-hTRM9L (dark grey bars) expressing cells were grown under 21% oxygen (normoxic) and 1% oxygen (hypoxic) conditions and the colonies formed were quantitated. No apparent growth difference on the cell lines was detected when grown under normal oxygen conditions, whereas exposure to hypoxia revealed a growth-sensitive phenotype of hTRM9L expressing cells.
C. Protein lysates prepared from 3-day-old tumours were added to MSD MULTI-SPOT 4-Spot plates coated with anti-total-HIF1-α antibody. Total HIF1-α was detected with anti-total-HIF1-α antibody labeled with MSD SULFO-TAG reagent. hTRM9L expressing tumour cells show a twofold increase in HIF1-α protein levels compared to control tumour cells.
D. Immunofluorescence microscopy analysis of HIF1-α in hTRM9L deficient and proficient cells under normoxic and hypoxic conditions. Images were not fixed in exposure time, thus, they do not reflect the quantitative protein abundance difference shown in (C).
E. Levels of downstream markers for HIF1-α target activation, GLUT1, were analysed in both cells types.
Complementation analysis of trm9Δ yeast cells with hTRM9L, hALKBH8 and yTrm9.
Hierarchical clustering of tRNA modification data, with significance calculated relative to trm9Δ cells. tRNA modification increases were assigned a positive p-value, while decreases were assigned a negative p-value.
SW620-lacZ and SW620-hTRM9L cells were left untreated or exposed to 1.5 mg/ml paromomycin for 24 h. For data reported in (B) and (C), small RNA was isolated and validated to be of high integrity by Bioanalyser analysis. RNA was enzymatically digested to nucleosides and the identity and levels, as determined by total ion count (TIC), of specific modified nucleosides was determined by LS/MS-MS analysis.
SW480 and SW620 cells were treated with paromomycin and the percent viability for each was determined by trypan blue staining 24 h post exposure.
SW620-LacZ and SW620-hTRM9L cells were exposed to paromomycin and assays were performed as described above.
Similar articles
-
Ovarian cancer proliferation and apoptosis are regulated by human transfer RNA methyltransferase 9-likevia LIN9.Oncol Lett. 2017 Oct;14(4):4461-4466. doi: 10.3892/ol.2017.6750. Epub 2017 Aug 14. Oncol Lett. 2017. PMID: 29085442 Free PMC article.
-
Silencing of KIAA1429, a N6-methyladenine methyltransferase, inhibits the progression of colon adenocarcinoma via blocking the hypoxia-inducible factor 1 signalling pathway.J Biochem Mol Toxicol. 2024 Sep;38(9):e23829. doi: 10.1002/jbt.23829. J Biochem Mol Toxicol. 2024. PMID: 39215765
-
RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated.Cancer Lett. 2016 Jun 28;376(1):34-42. doi: 10.1016/j.canlet.2016.02.022. Epub 2016 Mar 17. Cancer Lett. 2016. PMID: 26996300
-
Hypoxia-inducible factor 1-alpha up-regulates the expression of phospholipase D2 in colon cancer cells under hypoxic conditions.Med Oncol. 2015 Jan;32(1):394. doi: 10.1007/s12032-014-0394-9. Epub 2014 Nov 29. Med Oncol. 2015. PMID: 25432699
-
Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification.RNA Biol. 2014;11(12):1608-18. doi: 10.1080/15476286.2015.1008360. RNA Biol. 2014. PMID: 25625329 Free PMC article. Review.
Cited by
-
Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation.PLoS Genet. 2015 Dec 15;11(12):e1005706. doi: 10.1371/journal.pgen.1005706. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26670883 Free PMC article.
-
Ovarian cancer proliferation and apoptosis are regulated by human transfer RNA methyltransferase 9-likevia LIN9.Oncol Lett. 2017 Oct;14(4):4461-4466. doi: 10.3892/ol.2017.6750. Epub 2017 Aug 14. Oncol Lett. 2017. PMID: 29085442 Free PMC article.
-
Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.Proc Natl Acad Sci U S A. 2015 May 12;112(19):6033-7. doi: 10.1073/pnas.1500161112. Epub 2015 Apr 22. Proc Natl Acad Sci U S A. 2015. PMID: 25902496 Free PMC article.
-
RNA modification: mechanisms and therapeutic targets.Mol Biomed. 2023 Aug 24;4(1):25. doi: 10.1186/s43556-023-00139-x. Mol Biomed. 2023. PMID: 37612540 Free PMC article. Review.
-
tRNA modifications regulate translation during cellular stress.FEBS Lett. 2014 Nov 28;588(23):4287-96. doi: 10.1016/j.febslet.2014.09.038. Epub 2014 Oct 7. FEBS Lett. 2014. PMID: 25304425 Free PMC article. Review.
References
-
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Recovery pathways in S. cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. - PubMed
-
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol Cell. 2004;16:117–125. - PubMed
-
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

