Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis
- PMID: 23354436
- PMCID: PMC3647333
- DOI: 10.1038/ng.2531
Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis
Erratum in
- Nat Genet. 2013 Oct;45(10):1261
Abstract
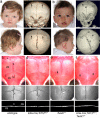
Craniosynostosis, the premature fusion of the cranial sutures, is a heterogeneous disorder with a prevalence of ∼1 in 2,200 (refs. 1,2). A specific genetic etiology can be identified in ∼21% of cases, including mutations of TWIST1, which encodes a class II basic helix-loop-helix (bHLH) transcription factor, and causes Saethre-Chotzen syndrome, typically associated with coronal synostosis. Using exome sequencing, we identified 38 heterozygous TCF12 mutations in 347 samples from unrelated individuals with craniosynostosis. The mutations predominantly occurred in individuals with coronal synostosis and accounted for 32% and 10% of subjects with bilateral and unilateral pathology, respectively. TCF12 encodes one of three class I E proteins that heterodimerize with class II bHLH proteins such as TWIST1. We show that TCF12 and TWIST1 act synergistically in a transactivation assay and that mice doubly heterozygous for loss-of-function mutations in Tcf12 and Twist1 have severe coronal synostosis. Hence, the dosage of TCF12-TWIST1 heterodimers is critical for normal coronal suture development.
Figures
Similar articles
-
Altered bone growth dynamics prefigure craniosynostosis in a zebrafish model of Saethre-Chotzen syndrome.Elife. 2018 Oct 25;7:e37024. doi: 10.7554/eLife.37024. Elife. 2018. PMID: 30375332 Free PMC article.
-
Clinical spectrum and outcomes in families with coronal synostosis and TCF12 mutations.Eur J Hum Genet. 2014 Dec;22(12):1413-6. doi: 10.1038/ejhg.2014.57. Epub 2014 Apr 16. Eur J Hum Genet. 2014. PMID: 24736737 Free PMC article.
-
Twist1 dimer selection regulates cranial suture patterning and fusion.Dev Dyn. 2006 May;235(5):1345-57. doi: 10.1002/dvdy.20717. Dev Dyn. 2006. PMID: 16502419
-
Phosphoregulation of Twist1 provides a mechanism of cell fate control.Curr Med Chem. 2008;15(25):2641-7. doi: 10.2174/092986708785908987. Curr Med Chem. 2008. PMID: 18855684 Free PMC article. Review.
-
Reoperation for intracranial hypertension in TWIST1-confirmed Saethre-Chotzen syndrome: a 15-year review.Plast Reconstr Surg. 2009 Jun;123(6):1801-1810. doi: 10.1097/PRS.0b013e3181a3f391. Plast Reconstr Surg. 2009. PMID: 19483581 Free PMC article. Review.
Cited by
-
Language Impairments in ASD Resulting from a Failed Domestication of the Human Brain.Front Neurosci. 2016 Aug 29;10:373. doi: 10.3389/fnins.2016.00373. eCollection 2016. Front Neurosci. 2016. PMID: 27621700 Free PMC article.
-
Co-occurrence of frameshift mutations in SMAD6 and TCF12 in a child with complex craniosynostosis.Hum Genome Var. 2018 Jun 28;5:14. doi: 10.1038/s41439-018-0014-x. eCollection 2018. Hum Genome Var. 2018. PMID: 30038786 Free PMC article.
-
Disruption of TWIST1 translation by 5' UTR variants in Saethre-Chotzen syndrome.Hum Mutat. 2018 Oct;39(10):1360-1365. doi: 10.1002/humu.23598. Epub 2018 Aug 7. Hum Mutat. 2018. PMID: 30040876 Free PMC article.
-
ERF-related craniosynostosis: The phenotypic and developmental profile of a new craniosynostosis syndrome.Am J Med Genet A. 2019 Apr;179(4):615-627. doi: 10.1002/ajmg.a.61073. Epub 2019 Feb 13. Am J Med Genet A. 2019. PMID: 30758909 Free PMC article.
-
Adapting SureSelect enrichment protocol to the Ion Torrent S5 platform in molecular diagnostics of craniosynostosis.Sci Rep. 2020 Mar 5;10(1):4159. doi: 10.1038/s41598-020-61048-5. Sci Rep. 2020. PMID: 32139749 Free PMC article.
References
-
- Lajeunie E, Le Merrer M, Bonaïti-Pellie C, Marchac D, Renier D. Genetic study of nonsyndromic coronal craniosynostosis. Am. J. Med. Genet. 1995;55:500–504. - PubMed
-
- Boulet SL, Rasmussen SA, Honein MA. A population-based study of craniosynostosis in metropolitan Atlanta, 1989-2003. Am. J. Med. Genet. 2008;146A:984–991. - PubMed
-
- Howard TD, et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat. Genet. 1997;15:36–41. - PubMed
-
- El Ghouzzi V, et al. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat. Genet. 1997;15:42–46. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

