PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis
- PMID: 23340338
- PMCID: PMC3683295
- DOI: 10.1038/nchembio.1166
PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis
Abstract
Following DNA damage, nuclear p53 induces the expression of PUMA, a BH3-only protein that binds and inhibits the antiapoptotic BCL-2 repertoire, including BCL-xL. PUMA, unique among BH3-only proteins, disrupts the interaction between cytosolic p53 and BCL-xL, allowing p53 to promote apoptosis via direct activation of the BCL-2 effector molecules BAX and BAK. Structural investigations using NMR spectroscopy and X-ray crystallography revealed that PUMA binding induced partial unfolding of two α-helices within BCL-xL. Wild-type PUMA or a PUMA mutant incapable of causing binding-induced unfolding of BCL-xL equivalently inhibited the antiapoptotic BCL-2 repertoire to sensitize for death receptor-activated apoptosis, but only wild-type PUMA promoted p53-dependent, DNA damage-induced apoptosis. Our data suggest that PUMA-induced partial unfolding of BCL-xL disrupts interactions between cytosolic p53 and BCL-xL, releasing the bound p53 to initiate apoptosis. We propose that regulated unfolding of BCL-xL provides a mechanism to promote PUMA-dependent signaling within the apoptotic pathways.
Figures
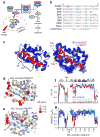
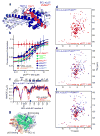

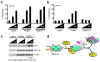
Comment in
-
Protein-protein interactions: A PUMA mechanism unfolds.Nat Chem Biol. 2013 Mar;9(3):141-3. doi: 10.1038/nchembio.1177. Nat Chem Biol. 2013. PMID: 23416398 Free PMC article.
Similar articles
-
Bax/Bak activation in the absence of Bid, Bim, Puma, and p53.Cell Death Dis. 2016 Jun 16;7(6):e2266. doi: 10.1038/cddis.2016.167. Cell Death Dis. 2016. PMID: 27310874 Free PMC article.
-
Development of small-molecule PUMA inhibitors for mitigating radiation-induced cell death.Curr Top Med Chem. 2011;11(3):281-90. doi: 10.2174/156802611794072641. Curr Top Med Chem. 2011. PMID: 21320058 Free PMC article.
-
p53 up-regulated modulator of apoptosis (PUMA) activation contributes to pancreatic beta-cell apoptosis induced by proinflammatory cytokines and endoplasmic reticulum stress.J Biol Chem. 2010 Jun 25;285(26):19910-20. doi: 10.1074/jbc.M110.122374. Epub 2010 Apr 26. J Biol Chem. 2010. PMID: 20421300 Free PMC article.
-
PUMA, a critical mediator of cell death--one decade on from its discovery.Cell Mol Biol Lett. 2012 Dec;17(4):646-69. doi: 10.2478/s11658-012-0032-5. Epub 2012 Sep 20. Cell Mol Biol Lett. 2012. PMID: 23001513 Free PMC article. Review.
-
PUMA, a potent killer with or without p53.Oncogene. 2008 Dec;27 Suppl 1(Suppl 1):S71-83. doi: 10.1038/onc.2009.45. Oncogene. 2008. PMID: 19641508 Free PMC article. Review.
Cited by
-
Anticancer compound ABT-263 accelerates apoptosis in virus-infected cells and imbalances cytokine production and lowers survival rates of infected mice.Cell Death Dis. 2013 Jul 25;4(7):e742. doi: 10.1038/cddis.2013.267. Cell Death Dis. 2013. PMID: 23887633 Free PMC article.
-
One-step cell biomanufacturing platform: porous gelatin microcarrier beads promote human embryonic stem cell-derived midbrain dopaminergic progenitor cell differentiation in vitro and survival after transplantation in vivo.Neural Regen Res. 2024 Feb;19(2):458-464. doi: 10.4103/1673-5374.377412. Neural Regen Res. 2024. PMID: 37488911 Free PMC article.
-
The DNA-binding domain mediates both nuclear and cytosolic functions of p53.Nat Struct Mol Biol. 2014 Jun;21(6):535-43. doi: 10.1038/nsmb.2829. Epub 2014 May 11. Nat Struct Mol Biol. 2014. PMID: 24814347 Free PMC article.
-
Computational approaches for inferring the functions of intrinsically disordered proteins.Front Mol Biosci. 2015 Aug 5;2:45. doi: 10.3389/fmolb.2015.00045. eCollection 2015. Front Mol Biosci. 2015. PMID: 26301226 Free PMC article. Review.
-
Structural basis for proapoptotic activation of Bak by the noncanonical BH3-only protein Pxt1.PLoS Biol. 2023 Jun 14;21(6):e3002156. doi: 10.1371/journal.pbio.3002156. eCollection 2023 Jun. PLoS Biol. 2023. PMID: 37315086 Free PMC article.
References
-
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. - PubMed
-
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. - PubMed
-
- Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. - PubMed
-
- Jeffers JR, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

