CENP-T provides a structural platform for outer kinetochore assembly
- PMID: 23334297
- PMCID: PMC3567495
- DOI: 10.1038/emboj.2012.348
CENP-T provides a structural platform for outer kinetochore assembly
Abstract
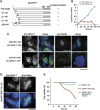
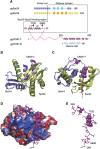
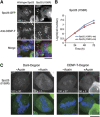
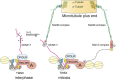
The kinetochore forms a dynamic interface with microtubules from the mitotic spindle during mitosis. The Ndc80 complex acts as the key microtubule-binding complex at kinetochores. However, it is unclear how the Ndc80 complex associates with the inner kinetochore proteins that assemble upon centromeric chromatin. Here, based on a high-resolution structural analysis, we demonstrate that the N-terminal region of vertebrate CENP-T interacts with the 'RWD' domain in the Spc24/25 portion of the Ndc80 complex. Phosphorylation of CENP-T strengthens a cryptic hydrophobic interaction between CENP-T and Spc25 resulting in a phospho-regulated interaction that occurs without direct recognition of the phosphorylated residue. The Ndc80 complex interacts with both CENP-T and the Mis12 complex, but we find that these interactions are mutually exclusive, supporting a model in which two distinct pathways target the Ndc80 complex to kinetochores. Our results provide a model for how the multiple protein complexes at kinetochores associate in a phospho-regulated manner.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T.Curr Biol. 2015 Mar 2;25(5):671-7. doi: 10.1016/j.cub.2015.01.059. Epub 2015 Feb 5. Curr Biol. 2015. PMID: 25660545 Free PMC article.
-
A quantitative description of Ndc80 complex linkage to human kinetochores.Nat Commun. 2015 Sep 8;6:8161. doi: 10.1038/ncomms9161. Nat Commun. 2015. PMID: 26345214 Free PMC article.
-
Molecular basis of outer kinetochore assembly on CENP-T.Elife. 2016 Dec 24;5:e21007. doi: 10.7554/eLife.21007. Elife. 2016. PMID: 28012276 Free PMC article.
-
The Ndc80 complex: hub of kinetochore activity.FEBS Lett. 2007 Jun 19;581(15):2862-9. doi: 10.1016/j.febslet.2007.05.012. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17521635 Review.
-
The ABCs of CENPs.Chromosoma. 2011 Oct;120(5):425-46. doi: 10.1007/s00412-011-0330-0. Epub 2011 Jul 13. Chromosoma. 2011. PMID: 21751032 Review.
Cited by
-
Plk1 protects kinetochore-centromere architecture against microtubule pulling forces.EMBO Rep. 2019 Oct 4;20(10):e48711. doi: 10.15252/embr.201948711. Epub 2019 Aug 30. EMBO Rep. 2019. PMID: 31468671 Free PMC article.
-
A CENP-S/X complex assembles at the centromere in S and G2 phases of the human cell cycle.Open Biol. 2014 Feb 12;4(2):130229. doi: 10.1098/rsob.130229. Open Biol. 2014. PMID: 24522885 Free PMC article.
-
Physicochemical mechanisms of protein regulation by phosphorylation.Front Genet. 2014 Aug 7;5:270. doi: 10.3389/fgene.2014.00270. eCollection 2014. Front Genet. 2014. PMID: 25147561 Free PMC article. Review.
-
Kinetochore biorientation in Saccharomyces cerevisiae requires a tightly folded conformation of the Ndc80 complex.Genetics. 2014 Dec;198(4):1483-93. doi: 10.1534/genetics.114.167775. Epub 2014 Sep 16. Genetics. 2014. PMID: 25230952 Free PMC article.
-
Immunization with CENP-C Causes Aberrant Chromosome Segregation during Oocyte Meiosis in Mice.J Immunol Res. 2021 Jan 30;2021:4610494. doi: 10.1155/2021/4610494. eCollection 2021. J Immunol Res. 2021. PMID: 33604391 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echools N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr D66: 213–221 - PMC - PubMed
-
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

