RNA-Seq analysis reveals new gene models and alternative splicing in the fungal pathogen Fusarium graminearum
- PMID: 23324402
- PMCID: PMC3577648
- DOI: 10.1186/1471-2164-14-21
RNA-Seq analysis reveals new gene models and alternative splicing in the fungal pathogen Fusarium graminearum
Abstract
Background: The genome of Fusarium graminearum has been sequenced and annotated previously, but correct gene annotation remains a challenge. In addition, posttranscriptional regulations, such as alternative splicing and RNA editing, are poorly understood in F. graminearum. Here we took advantage of RNA-Seq to improve gene annotations and to identify alternative splicing and RNA editing in F. graminearum.
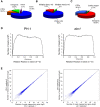
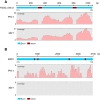
Results: We identified and revised 655 incorrectly predicted gene models, including revisions of intron predictions, intron splice sites and prediction of novel introns. 231 genes were identified with two or more alternative splice variants, mostly due to intron retention. Interestingly, the expression ratios between different transcript isoforms appeared to be developmentally regulated. Surprisingly, no RNA editing was identified in F. graminearum. Moreover, 2459 novel transcriptionally active regions (nTARs) were identified and our analysis indicates that many of these could be missed genes. Finally, we identified the 5' UTR and/or 3' UTR sequences of 7666 genes. A number of representative novel gene models and alternatively spliced genes were validated by reverse transcription polymerase chain reaction and sequencing of the generated amplicons.
Conclusions: We have developed novel and efficient strategies to identify alternatively spliced genes and incorrect gene models based on RNA-Seq data. Our study identified hundreds of alternatively spliced genes in F. graminearum and for the first time indicated that alternative splicing is developmentally regulated in filamentous fungi. In addition, hundreds of incorrect predicted gene models were identified and revised and thousands of nTARs were discovered in our study, which will be helpful for the future genomic and transcriptomic studies in F. graminearum.
Figures


Similar articles
-
New gene models and alternative splicing in the maize pathogen Colletotrichum graminicola revealed by RNA-Seq analysis.BMC Genomics. 2014 Oct 2;15(1):842. doi: 10.1186/1471-2164-15-842. BMC Genomics. 2014. PMID: 25281481 Free PMC article.
-
The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum.BMC Genomics. 2015 Jul 22;16(1):544. doi: 10.1186/s12864-015-1756-1. BMC Genomics. 2015. PMID: 26198851 Free PMC article.
-
The presence of GC-AG introns in Neurospora crassa and other euascomycetes determined from analyses of complete genomes: implications for automated gene prediction.Genomics. 2006 Mar;87(3):338-47. doi: 10.1016/j.ygeno.2005.11.014. Epub 2006 Jan 9. Genomics. 2006. PMID: 16406724
-
How did alternative splicing evolve?Nat Rev Genet. 2004 Oct;5(10):773-82. doi: 10.1038/nrg1451. Nat Rev Genet. 2004. PMID: 15510168 Review.
-
Multiplexed primer extension sequencing: A targeted RNA-seq method that enables high-precision quantitation of mRNA splicing isoforms and rare pre-mRNA splicing intermediates.Methods. 2020 Apr 1;176:34-45. doi: 10.1016/j.ymeth.2019.05.013. Epub 2019 May 21. Methods. 2020. PMID: 31121301 Free PMC article. Review.
Cited by
-
Unanticipated Large-Scale Deletion in Fusarium graminearum Genome Using CRISPR/Cas9 and Its Impact on Growth and Virulence.J Fungi (Basel). 2023 Jun 14;9(6):673. doi: 10.3390/jof9060673. J Fungi (Basel). 2023. PMID: 37367609 Free PMC article.
-
Lessons on fruiting body morphogenesis from genomes and transcriptomes of Agaricomycetes.Stud Mycol. 2023 Jul;104:1-85. doi: 10.3114/sim.2022.104.01. Epub 2023 Jan 31. Stud Mycol. 2023. PMID: 37351542 Free PMC article.
-
New insights into genome annotation in Podospora anserina through re-exploiting multiple RNA-seq data.BMC Genomics. 2022 Dec 29;23(1):859. doi: 10.1186/s12864-022-09085-4. BMC Genomics. 2022. PMID: 36581831 Free PMC article.
-
Relation between CarS expression and activation of carotenogenesis by stress in Fusarium fujikuroi.Front Bioeng Biotechnol. 2022 Oct 5;10:1000129. doi: 10.3389/fbioe.2022.1000129. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36277400 Free PMC article.
-
TuRLK1, a leucine-rich repeat receptor-like kinase, is indispensable for stripe rust resistance of YrU1 and confers broad resistance to multiple pathogens.BMC Plant Biol. 2022 Jun 8;22(1):280. doi: 10.1186/s12870-022-03679-6. BMC Plant Biol. 2022. PMID: 35676630 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

