Deciphering signatures of mutational processes operative in human cancer
- PMID: 23318258
- PMCID: PMC3588146
- DOI: 10.1016/j.celrep.2012.12.008
Deciphering signatures of mutational processes operative in human cancer
Abstract
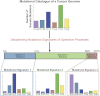
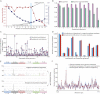
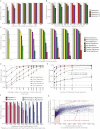
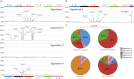
The genome of a cancer cell carries somatic mutations that are the cumulative consequences of the DNA damage and repair processes operative during the cellular lineage between the fertilized egg and the cancer cell. Remarkably, these mutational processes are poorly characterized. Global sequencing initiatives are yielding catalogs of somatic mutations from thousands of cancers, thus providing the unique opportunity to decipher the signatures of mutational processes operative in human cancer. However, until now there have been no theoretical models describing the signatures of mutational processes operative in cancer genomes and no systematic computational approaches are available to decipher these mutational signatures. Here, by modeling mutational processes as a blind source separation problem, we introduce a computational framework that effectively addresses these questions. Our approach provides a basis for characterizing mutational signatures from cancer-derived somatic mutational catalogs, paving the way to insights into the pathogenetic mechanism underlying all cancers.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
The topography of mutational processes in breast cancer genomes.Nat Commun. 2016 May 2;7:11383. doi: 10.1038/ncomms11383. Nat Commun. 2016. PMID: 27136393 Free PMC article.
-
The repertoire of mutational signatures in human cancer.Nature. 2020 Feb;578(7793):94-101. doi: 10.1038/s41586-020-1943-3. Epub 2020 Feb 5. Nature. 2020. PMID: 32025018 Free PMC article.
-
Mutational spectra and mutational signatures: Insights into cancer aetiology and mechanisms of DNA damage and repair.DNA Repair (Amst). 2018 Nov;71:6-11. doi: 10.1016/j.dnarep.2018.08.003. Epub 2018 Aug 24. DNA Repair (Amst). 2018. PMID: 30236628 Free PMC article. Review.
-
Bioinformatic Methods to Identify Mutational Signatures in Cancer.Methods Mol Biol. 2021;2185:447-473. doi: 10.1007/978-1-0716-0810-4_28. Methods Mol Biol. 2021. PMID: 33165866
-
Mechanisms underlying mutational signatures in human cancers.Nat Rev Genet. 2014 Sep;15(9):585-98. doi: 10.1038/nrg3729. Epub 2014 Jul 1. Nat Rev Genet. 2014. PMID: 24981601 Free PMC article. Review.
Cited by
-
Cytidine Deaminase APOBEC3A Regulates PD-L1 Expression in Cancer Cells in a JNK/c-JUN-Dependent Manner.Mol Cancer Res. 2021 Sep;19(9):1571-1582. doi: 10.1158/1541-7786.MCR-21-0219. Epub 2021 May 27. Mol Cancer Res. 2021. PMID: 34045311 Free PMC article.
-
The topography of mutational processes in breast cancer genomes.Nat Commun. 2016 May 2;7:11383. doi: 10.1038/ncomms11383. Nat Commun. 2016. PMID: 27136393 Free PMC article.
-
Genome-wide mutational spectra analysis reveals significant cancer-specific heterogeneity.Sci Rep. 2015 Jul 27;5:12566. doi: 10.1038/srep12566. Sci Rep. 2015. PMID: 26212640 Free PMC article.
-
Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors.Nat Genet. 2016 Nov;48(11):1330-1338. doi: 10.1038/ng.3670. Epub 2016 Sep 19. Nat Genet. 2016. PMID: 27643540 Free PMC article.
-
Tumour mutational burden: clinical utility, challenges and emerging improvements.Nat Rev Clin Oncol. 2024 Oct;21(10):725-742. doi: 10.1038/s41571-024-00932-9. Epub 2024 Aug 27. Nat Rev Clin Oncol. 2024. PMID: 39192001 Review.
References
-
- Ames B.N., Gold L.S. Endogenous mutagens and the causes of aging and cancer. Mutat. Res. 1991;250:3–16. - PubMed
-
- Berry M.W., Browne M., Langville A.N., Pauca V.P., Plemmons R.J. Algorithms and applications for approximate nonnegative matrix factorization. Comput. Stat. Data Anal. 2007;52:155–173.
-
- Berwick M., Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J. Natl. Cancer Inst. 2000;92:874–897. - PubMed
-
- Comon P. First Edition. Elsevier; Boston, MA: 2010. Handbook of Blind Source Separation: Independent Component Analysis and Blind Deconvolution.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

