NRP1 acts cell autonomously in endothelium to promote tip cell function during sprouting angiogenesis
- PMID: 23315162
- PMCID: PMC3606070
- DOI: 10.1182/blood-2012-05-424713
NRP1 acts cell autonomously in endothelium to promote tip cell function during sprouting angiogenesis
Abstract
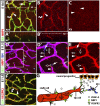
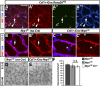
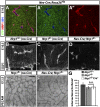
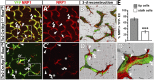
Neuropilin (NRP) 1 is a receptor for the vascular endothelial growth factor (VEGF)-A and is essential for normal angiogenesis. Previous in vitro experiments identified NRP1 interactions with VEGF-A's main signaling receptor VEGFR2 within endothelial cells, but also between nonendothelial NRP1 and endothelial VEGFR2. Consistent with an endothelial role for NRP1 in angiogenesis, we found that VEGFR2 and NRP1 were coexpressed in endothelial tip and stalk cells in the developing brain. In addition, NRP1 was expressed on two cell types that interact with growing brain vessels-the neural progenitors that secrete VEGF-A to stimulate tip cell activity and the pro-angiogenic macrophages that promote tip cell anastomosis. Selective targeting of Nrp1 in each of these cell types demonstrated that neural progenitor- and macrophage-derived NRP1 were dispensable, whereas endothelial NRP1 was essential for normal brain vessel growth. NRP1 therefore promotes brain angiogenesis cell autonomously in endothelium, independently of heterotypic interactions with nonendothelial cells. Genetic mosaic analyses demonstrated a key role for NRP1 in endothelial tip rather than stalk cells during vessel sprouting. Thus, NRP1-expressing endothelial cells attained the tip cell position when competing with NRP1-negative endothelial cells in chimeric vessel sprouts. Taken together, these findings demonstrate that NRP1 promotes endothelial tip cell function during angiogenesis.
Figures


Similar articles
-
NRP1 Regulates CDC42 Activation to Promote Filopodia Formation in Endothelial Tip Cells.Cell Rep. 2015 Jun 16;11(10):1577-90. doi: 10.1016/j.celrep.2015.05.018. Epub 2015 Jun 4. Cell Rep. 2015. PMID: 26051942 Free PMC article.
-
VEGF binding to NRP1 is essential for VEGF stimulation of endothelial cell migration, complex formation between NRP1 and VEGFR2, and signaling via FAK Tyr407 phosphorylation.Mol Biol Cell. 2011 Aug 1;22(15):2766-76. doi: 10.1091/mbc.E09-12-1061. Epub 2011 Jun 8. Mol Biol Cell. 2011. PMID: 21653826 Free PMC article.
-
Neuropilin-1 binds to VEGF121 and regulates endothelial cell migration and sprouting.J Biol Chem. 2007 Aug 17;282(33):24049-56. doi: 10.1074/jbc.M703554200. Epub 2007 Jun 16. J Biol Chem. 2007. PMID: 17575273
-
Neuropilin-1 enforces extracellular matrix signalling via ABL1 to promote angiogenesis.Biochem Soc Trans. 2014 Oct;42(5):1429-34. doi: 10.1042/BST20140141. Biochem Soc Trans. 2014. PMID: 25233427 Review.
-
The role of neuropilin in vascular and tumor biology.Adv Exp Med Biol. 2002;515:33-48. doi: 10.1007/978-1-4615-0119-0_3. Adv Exp Med Biol. 2002. PMID: 12613541 Review.
Cited by
-
ELAVL1 modulates transcriptome-wide miRNA binding in murine macrophages.Cell Rep. 2014 Dec 24;9(6):2330-43. doi: 10.1016/j.celrep.2014.11.030. Epub 2014 Dec 18. Cell Rep. 2014. PMID: 25533351 Free PMC article.
-
Proteomic analysis of bone marrow-derived mesenchymal stem cell extracellular vesicles from healthy donors: implications for proliferation, angiogenesis, Wnt signaling, and the basement membrane.Stem Cell Res Ther. 2021 Jun 5;12(1):328. doi: 10.1186/s13287-021-02405-7. Stem Cell Res Ther. 2021. PMID: 34090527 Free PMC article.
-
Quantifying and Characterizing Angiogenesis Using the Postnatal Mouse Retina.Methods Mol Biol. 2022;2441:63-73. doi: 10.1007/978-1-0716-2059-5_5. Methods Mol Biol. 2022. PMID: 35099728
-
Knockdown of HIF-1α impairs post-ischemic vascular reconstruction in the brain via deficient homing and sprouting bmEPCs.Brain Pathol. 2018 Nov;28(6):860-874. doi: 10.1111/bpa.12628. Epub 2018 Oct 9. Brain Pathol. 2018. PMID: 30052311 Free PMC article.
-
Neuropilin-1 deficiency in vascular smooth muscle cells is associated with hereditary hemorrhagic telangiectasia arteriovenous malformations.JCI Insight. 2022 May 9;7(9):e155565. doi: 10.1172/jci.insight.155565. JCI Insight. 2022. PMID: 35380991 Free PMC article.
References
-
- Ruhrberg C. Growing and shaping the vascular tree: multiple roles for VEGF. BioEssays. 2003;25(11):1052–1060. - PubMed
-
- Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438(7070):932–936. - PubMed
-
- Koch S, Tugues S, Li X, Gualandi L, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Biochem J. 2011;437(2):169–183. - PubMed
-
- Shalaby F, Rossant J, Yamaguchi TP, et al. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376(6535):62–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

