Folding and binding of an intrinsically disordered protein: fast, but not 'diffusion-limited'
- PMID: 23301700
- PMCID: PMC3776562
- DOI: 10.1021/ja309527h
Folding and binding of an intrinsically disordered protein: fast, but not 'diffusion-limited'
Abstract
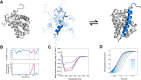
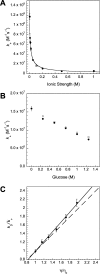
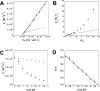
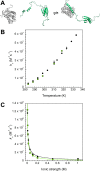
Coupled folding and binding of intrinsically disordered proteins (IDPs) is prevalent in biology. As the first step toward understanding the mechanism of binding, it is important to know if a reaction is 'diffusion-limited' as, if this speed limit is reached, the association must proceed through an induced fit mechanism. Here, we use a model system where the 'BH3 region' of PUMA, an IDP, forms a single, contiguous α-helix upon binding the folded protein Mcl-1. Using stopped-flow techniques, we systematically compare the rate constant for association (k(+)) under a number of solvent conditions and temperatures. We show that our system is not 'diffusion-limited', despite having a k(+) in the often-quoted 'diffusion-limited' regime (10(5)-10(6) M(-1) s(-1) at high ionic strength) and displaying an inverse dependence on solvent viscosity. These standard tests, developed for folded protein-protein interactions, are not appropriate for reactions where one protein is disordered.
Figures
Similar articles
-
Coupled folding and binding of the disordered protein PUMA does not require particular residual structure.J Am Chem Soc. 2014 Apr 9;136(14):5197-200. doi: 10.1021/ja4125065. Epub 2014 Mar 31. J Am Chem Soc. 2014. PMID: 24654952 Free PMC article.
-
Folding and binding pathways of BH3-only proteins are encoded within their intrinsically disordered sequence, not templated by partner proteins.J Biol Chem. 2018 Jun 22;293(25):9718-9723. doi: 10.1074/jbc.RA118.002791. Epub 2018 May 1. J Biol Chem. 2018. PMID: 29716994 Free PMC article.
-
Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding.Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):9614-9. doi: 10.1073/pnas.1512799112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195786 Free PMC article.
-
Uncoupled binding and folding of immune signaling-related intrinsically disordered proteins.Prog Biophys Mol Biol. 2011 Sep;106(3):525-36. doi: 10.1016/j.pbiomolbio.2011.08.005. Epub 2011 Aug 18. Prog Biophys Mol Biol. 2011. PMID: 21867726 Review.
-
Features of molecular recognition of intrinsically disordered proteins via coupled folding and binding.Protein Sci. 2019 Nov;28(11):1952-1965. doi: 10.1002/pro.3718. Epub 2019 Sep 4. Protein Sci. 2019. PMID: 31441158 Free PMC article. Review.
Cited by
-
Intrinsically Disordered Proteins: An Overview.Int J Mol Sci. 2022 Nov 14;23(22):14050. doi: 10.3390/ijms232214050. Int J Mol Sci. 2022. PMID: 36430530 Free PMC article. Review.
-
A disordered encounter complex is central to the yeast Abp1p SH3 domain binding pathway.PLoS Comput Biol. 2020 Sep 14;16(9):e1007815. doi: 10.1371/journal.pcbi.1007815. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32925900 Free PMC article.
-
Conformational entropy of a single peptide controlled under force governs protease recognition and catalysis.Proc Natl Acad Sci U S A. 2018 Nov 6;115(45):11525-11530. doi: 10.1073/pnas.1803872115. Epub 2018 Oct 19. Proc Natl Acad Sci U S A. 2018. PMID: 30341218 Free PMC article.
-
Conformational Ensembles Exhibit Extensive Molecular Recognition Features.ACS Omega. 2018 Aug 24;3(8):9907-9920. doi: 10.1021/acsomega.8b00898. eCollection 2018 Aug 31. ACS Omega. 2018. PMID: 31459119 Free PMC article.
-
Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs).Chem Rev. 2014 Jul 9;114(13):6661-714. doi: 10.1021/cr400695p. Epub 2014 Jun 5. Chem Rev. 2014. PMID: 24901537 Free PMC article. Review. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

