The circadian clock coordinates ribosome biogenesis
- PMID: 23300384
- PMCID: PMC3536797
- DOI: 10.1371/journal.pbio.1001455
The circadian clock coordinates ribosome biogenesis
Abstract
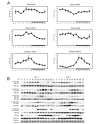
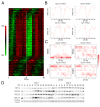
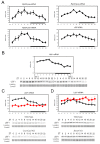
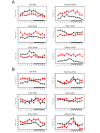
Biological rhythms play a fundamental role in the physiology and behavior of most living organisms. Rhythmic circadian expression of clock-controlled genes is orchestrated by a molecular clock that relies on interconnected negative feedback loops of transcription regulators. Here we show that the circadian clock exerts its function also through the regulation of mRNA translation. Namely, the circadian clock influences the temporal translation of a subset of mRNAs involved in ribosome biogenesis by controlling the transcription of translation initiation factors as well as the clock-dependent rhythmic activation of signaling pathways involved in their regulation. Moreover, the circadian oscillator directly regulates the transcription of ribosomal protein mRNAs and ribosomal RNAs. Thus the circadian clock exerts a major role in coordinating transcription and translation steps underlying ribosome biogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Comment in
-
Circadian genetics: Translation gets timely.Nat Rev Genet. 2013 Mar;14(3):153. doi: 10.1038/nrg3429. Epub 2013 Jan 22. Nat Rev Genet. 2013. PMID: 23338016 No abstract available.
Similar articles
-
Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver.Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):E6579-88. doi: 10.1073/pnas.1515308112. Epub 2015 Nov 9. Proc Natl Acad Sci U S A. 2015. PMID: 26554015 Free PMC article.
-
Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms.Proc Natl Acad Sci U S A. 2021 Jan 19;118(3):e2015803118. doi: 10.1073/pnas.2015803118. Proc Natl Acad Sci U S A. 2021. PMID: 33452134 Free PMC article.
-
Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames.Genome Res. 2015 Dec;25(12):1848-59. doi: 10.1101/gr.195404.115. Epub 2015 Oct 20. Genome Res. 2015. PMID: 26486724 Free PMC article.
-
mTORC1 signaling controls multiple steps in ribosome biogenesis.Semin Cell Dev Biol. 2014 Dec;36:113-20. doi: 10.1016/j.semcdb.2014.08.004. Epub 2014 Aug 19. Semin Cell Dev Biol. 2014. PMID: 25148809 Review.
-
The role of spatiotemporal organization and dynamics of clock complexes in circadian regulation.Curr Opin Cell Biol. 2022 Oct;78:102129. doi: 10.1016/j.ceb.2022.102129. Epub 2022 Sep 18. Curr Opin Cell Biol. 2022. PMID: 36126370 Free PMC article. Review.
Cited by
-
Nutrient-sensitive protein O-GlcNAcylation shapes daily biological rhythms.Open Biol. 2022 Sep;12(9):220215. doi: 10.1098/rsob.220215. Epub 2022 Sep 14. Open Biol. 2022. PMID: 36099933 Free PMC article. Review.
-
Regulation of protein O-GlcNAcylation by circadian, metabolic, and cellular signals.J Biol Chem. 2024 Feb;300(2):105616. doi: 10.1016/j.jbc.2023.105616. Epub 2023 Dec 29. J Biol Chem. 2024. PMID: 38159854 Free PMC article. Review.
-
The molecular clockwork of mammalian cells.Semin Cell Dev Biol. 2022 Jun;126:87-96. doi: 10.1016/j.semcdb.2021.03.012. Epub 2021 Mar 31. Semin Cell Dev Biol. 2022. PMID: 33810978 Free PMC article. Review.
-
A circadian clock translational control mechanism targets specific mRNAs to cytoplasmic messenger ribonucleoprotein granules.Cell Rep. 2022 Dec 27;41(13):111879. doi: 10.1016/j.celrep.2022.111879. Cell Rep. 2022. PMID: 36577368 Free PMC article.
-
Clocking In, Working Out: Circadian Regulation of Exercise Physiology.Trends Endocrinol Metab. 2019 Jun;30(6):347-356. doi: 10.1016/j.tem.2019.04.003. Epub 2019 May 2. Trends Endocrinol Metab. 2019. PMID: 31054802 Free PMC article. Review.
References
-
- Zhang EE, Kay SA (2010) Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776. - PubMed
-
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, et al. (2006) Circadian orchestration of the hepatic proteome. Curr Biol 16: 1107–1115. - PubMed
-
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. (2002) coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

