Diversion of stress granules and P-bodies during viral infection
- PMID: 23290869
- PMCID: PMC3611887
- DOI: 10.1016/j.virol.2012.11.017
Diversion of stress granules and P-bodies during viral infection
Abstract
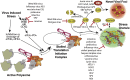
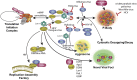
RNA granules are structures within cells that impart key regulatory measures on gene expression. Two general types of RNA granules are conserved from yeast to mammals: stress granules (SGs), which contain many translation initiation factors, and processing bodies (P-bodies, PBs), which are enriched for proteins involved in RNA turnover. Because of the inverse relationship between appearance of RNA granules and persistence of translation, many viruses must subvert RNA granule function for replicative purposes. Here we discuss the viruses and mechanisms that manipulate stress granules and P-bodies to promote synthesis of viral proteins. Several themes have emerged for manipulation of RNA granules by viruses: (1) disruption of RNA granules at the mid-phase of infection, (2) prevention of RNA granule assembly throughout infection and (3) co-opting of RNA granule proteins for new or parallel roles in viral reproduction. Viruses must employ one or multiple of these routes for a robust and productive infection to occur. The possible role for RNA granules in promoting innate immune responses poses an additional reason why viruses must counteract the effects of RNA granules for efficient replication.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Regulation of stress granules and P-bodies during RNA virus infection.Wiley Interdiscip Rev RNA. 2013 May-Jun;4(3):317-31. doi: 10.1002/wrna.1162. Epub 2013 Apr 3. Wiley Interdiscip Rev RNA. 2013. PMID: 23554219 Free PMC article. Review.
-
Who Regulates Whom? An Overview of RNA Granules and Viral Infections.Viruses. 2016 Jun 28;8(7):180. doi: 10.3390/v8070180. Viruses. 2016. PMID: 27367717 Free PMC article. Review.
-
Viral modulation of stress granules.Virus Res. 2012 Nov;169(2):430-7. doi: 10.1016/j.virusres.2012.06.004. Epub 2012 Jun 15. Virus Res. 2012. PMID: 22705970 Free PMC article. Review.
-
Cytoplasmic RNA Granules and Viral Infection.Annu Rev Virol. 2014 Nov;1(1):147-70. doi: 10.1146/annurev-virology-031413-085505. Annu Rev Virol. 2014. PMID: 26958719 Free PMC article.
-
Zika Virus Subverts Stress Granules To Promote and Restrict Viral Gene Expression.J Virol. 2019 May 29;93(12):e00520-19. doi: 10.1128/JVI.00520-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30944179 Free PMC article.
Cited by
-
Yin and yang regulation of stress granules by Caprin-1.Proc Natl Acad Sci U S A. 2022 Nov;119(44):e2207975119. doi: 10.1073/pnas.2207975119. Epub 2022 Oct 24. Proc Natl Acad Sci U S A. 2022. PMID: 36279435 Free PMC article.
-
Poly(ADP-ribose) polymerase-13 and RNA regulation in immunity and cancer.Trends Mol Med. 2015 Jun;21(6):373-84. doi: 10.1016/j.molmed.2015.03.002. Epub 2015 Apr 4. Trends Mol Med. 2015. PMID: 25851173 Free PMC article. Review.
-
Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation.PLoS Pathog. 2015 Feb 6;11(2):e1004659. doi: 10.1371/journal.ppat.1004659. eCollection 2015 Feb. PLoS Pathog. 2015. PMID: 25658430 Free PMC article.
-
Virus-induced translational arrest through 4EBP1/2-dependent decay of 5'-TOP mRNAs restricts viral infection.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):E2920-9. doi: 10.1073/pnas.1418805112. Epub 2015 May 18. Proc Natl Acad Sci U S A. 2015. PMID: 26038567 Free PMC article.
-
Principles and Properties of Stress Granules.Trends Cell Biol. 2016 Sep;26(9):668-679. doi: 10.1016/j.tcb.2016.05.004. Epub 2016 Jun 9. Trends Cell Biol. 2016. PMID: 27289443 Free PMC article. Review.
References
-
- Abrahamyan L.G., Chatel-Chaix L., Ajamian L., Milev M.P., Monette A., Clément J.-F., Song R., Lehmann M., DesGroseillers L., Laughrea M., Boccaccio G., Mouland A.J. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 2010;123:369–383. - PubMed
-
- Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. - PubMed
-
- Angus A.G.N., Dalrymple D., Boulant S., Mcgivern D.R., Clayton R.F., Scott M.J., Adair R., Graham S., Owsianka A.M., Targett-Adams P., Li K., Wakita T., McLauchlan J., Lemon S.M., Patel A.H. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J. Gen. Virol. 2010;91:122–132. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

