The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly
- PMID: 23277427
- PMCID: PMC3542802
- DOI: 10.1083/jcb.201210106
The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly
Abstract
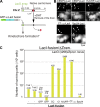
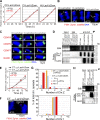
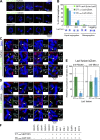
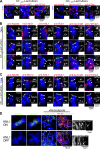
CENP-A acts as an important epigenetic marker for kinetochore specification. However, the mechanisms by which CENP-A is incorporated into centromeres and the structural basis for kinetochore formation downstream of CENP-A remain unclear. Here, we used a unique chromosome-engineering system in which kinetochore proteins are targeted to a noncentromeric site after the endogenous centromere is conditionally removed. Using this system, we created two distinct types of engineered kinetochores, both of which were stably maintained in chicken DT40 cells. Ectopic targeting of full-length HJURP, CENP-C, CENP-I, or the CENP-C C terminus generated engineered kinetochores containing major kinetochore components, including CENP-A. In contrast, ectopic targeting of the CENP-T or CENP-C N terminus generated functional kinetochores that recruit the microtubule-binding Ndc80 complex and chromosome passenger complex (CPC), but lack CENP-A and most constitutive centromere-associated network (CCAN) proteins. Based on the analysis of these different engineered kinetochores, we conclude that the CCAN has two distinct roles: recruiting CENP-A to establish the kinetochore and serving as a structural core to directly recruit kinetochore proteins.
Figures


Comment in
-
Reductionism at the vertebrate kinetochore.J Cell Biol. 2013 Jan 7;200(1):7-8. doi: 10.1083/jcb.201212005. Epub 2012 Dec 31. J Cell Biol. 2013. PMID: 23277428 Free PMC article.
Similar articles
-
CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore.Cell. 2008 Dec 12;135(6):1039-52. doi: 10.1016/j.cell.2008.10.019. Cell. 2008. PMID: 19070575
-
CENP-C and CENP-I are key connecting factors for kinetochore and CENP-A assembly.J Cell Sci. 2015 Dec 15;128(24):4572-87. doi: 10.1242/jcs.180786. Epub 2015 Nov 2. J Cell Sci. 2015. PMID: 26527398 Free PMC article.
-
HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore.J Cell Biol. 2011 Jul 25;194(2):229-43. doi: 10.1083/jcb.201012017. Epub 2011 Jul 18. J Cell Biol. 2011. PMID: 21768289 Free PMC article.
-
Conserved and divergent mechanisms of inner kinetochore assembly onto centromeric chromatin.Curr Opin Struct Biol. 2023 Aug;81:102638. doi: 10.1016/j.sbi.2023.102638. Epub 2023 Jun 20. Curr Opin Struct Biol. 2023. PMID: 37343495 Review.
-
The ABCs of CENPs.Chromosoma. 2011 Oct;120(5):425-46. doi: 10.1007/s00412-011-0330-0. Epub 2011 Jul 13. Chromosoma. 2011. PMID: 21751032 Review.
Cited by
-
Permitted and restricted steps of human kinetochore assembly in mitotic cell extracts.Mol Biol Cell. 2021 Jun 15;32(13):1241-1255. doi: 10.1091/mbc.E20-07-0461. Epub 2021 May 6. Mol Biol Cell. 2021. PMID: 33956511 Free PMC article.
-
Coregulation of NDC80 Complex Subunits Determines the Fidelity of the Spindle-Assembly Checkpoint and Mitosis.Mol Cancer Res. 2024 May 2;22(5):423-439. doi: 10.1158/1541-7786.MCR-23-0828. Mol Cancer Res. 2024. PMID: 38324016 Free PMC article.
-
Contractile acto-myosin network on nuclear envelope remnants positions human chromosomes for mitosis.Elife. 2019 Jul 3;8:e46902. doi: 10.7554/eLife.46902. Elife. 2019. PMID: 31264963 Free PMC article.
-
CENP-C/H/I/K/M/T/W/N/L and hMis12 but not CENP-S/X participate in complex formation in the nucleoplasm of living human interphase cells outside centromeres.PLoS One. 2018 Mar 6;13(3):e0192572. doi: 10.1371/journal.pone.0192572. eCollection 2018. PLoS One. 2018. PMID: 29509805 Free PMC article.
-
Cellular and Molecular Mechanisms of Centromere Drive.Cold Spring Harb Symp Quant Biol. 2017;82:249-257. doi: 10.1101/sqb.2017.82.034298. Epub 2018 Feb 12. Cold Spring Harb Symp Quant Biol. 2017. PMID: 29440567 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

