MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system
- PMID: 23260135
- PMCID: PMC3653293
- DOI: 10.1016/j.cell.2012.11.022
MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system
Abstract
The high level of 5-hydroxymethylcytosine (5hmC) present in neuronal genomes suggests that mechanisms interpreting 5hmC in the CNS may differ from those present in embryonic stem cells. Here, we present quantitative, genome-wide analysis of 5hmC, 5-methylcytosine (5mC), and gene expression in differentiated CNS cell types in vivo. We report that 5hmC is enriched in active genes and that, surprisingly, strong depletion of 5mC is observed over these regions. The contribution of these epigenetic marks to gene expression depends critically on cell type. We identify methyl-CpG-binding protein 2 (MeCP2) as the major 5hmC-binding protein in the brain and demonstrate that MeCP2 binds 5hmC- and 5mC-containing DNA with similar high affinities. The Rett-syndrome-causing mutation R133C preferentially inhibits 5hmC binding. These findings support a model in which 5hmC and MeCP2 constitute a cell-specific epigenetic mechanism for regulation of chromatin structure and gene expression.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
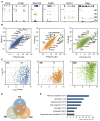
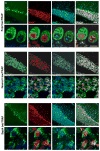
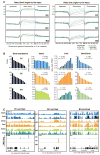
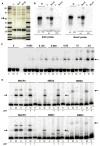
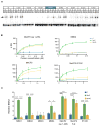
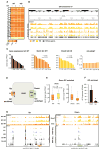
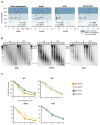
Comment in
-
By hook or by crook: multifaceted DNA-binding properties of MeCP2.Cell. 2013 Feb 28;152(5):940-2. doi: 10.1016/j.cell.2013.02.017. Cell. 2013. PMID: 23452844 Free PMC article.
Similar articles
-
Association of modified cytosines and the methylated DNA-binding protein MeCP2 with distinctive structural domains of lampbrush chromatin.Chromosome Res. 2012 Dec;20(8):925-42. doi: 10.1007/s10577-012-9324-x. Chromosome Res. 2012. PMID: 23149574 Free PMC article.
-
MeCP2 is a microsatellite binding protein that protects CA repeats from nucleosome invasion.Science. 2021 Jun 25;372(6549):eabd5581. doi: 10.1126/science.abd5581. Science. 2021. PMID: 34324427
-
5-Hydroxymethylcytosine-mediated active demethylation is required for mammalian neuronal differentiation and function.Elife. 2021 Dec 17;10:e66973. doi: 10.7554/eLife.66973. Elife. 2021. PMID: 34919053 Free PMC article.
-
Genomic distribution and possible functions of DNA hydroxymethylation in the brain.Genomics. 2014 Nov;104(5):341-6. doi: 10.1016/j.ygeno.2014.08.020. Epub 2014 Sep 7. Genomics. 2014. PMID: 25205307 Review.
-
The role of MeCP2 in CNS development and function.Horm Behav. 2011 Mar;59(3):364-8. doi: 10.1016/j.yhbeh.2010.05.014. Epub 2010 May 31. Horm Behav. 2011. PMID: 20515694 Free PMC article. Review.
Cited by
-
Epigenetic layers and players underlying neurodevelopment.Trends Neurosci. 2013 Aug;36(8):460-70. doi: 10.1016/j.tins.2013.05.001. Epub 2013 May 31. Trends Neurosci. 2013. PMID: 23731492 Free PMC article. Review.
-
The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases.Front Neurosci. 2014 Dec 5;8:397. doi: 10.3389/fnins.2014.00397. eCollection 2014. Front Neurosci. 2014. PMID: 25538551 Free PMC article. Review.
-
LINE-1 transcription in round spermatids is associated with accretion of 5-carboxylcytosine in their open reading frames.Commun Biol. 2021 Jun 7;4(1):691. doi: 10.1038/s42003-021-02217-8. Commun Biol. 2021. PMID: 34099857 Free PMC article.
-
Binding Analysis of Methyl-CpG Binding Domain of MeCP2 and Rett Syndrome Mutations.ACS Chem Biol. 2016 Oct 21;11(10):2706-2715. doi: 10.1021/acschembio.6b00450. Epub 2016 Aug 8. ACS Chem Biol. 2016. PMID: 27356039 Free PMC article.
-
Distinctive Patterns of 5-Methylcytosine and 5-Hydroxymethylcytosine in Schizophrenia.Int J Mol Sci. 2024 Jan 4;25(1):636. doi: 10.3390/ijms25010636. Int J Mol Sci. 2024. PMID: 38203806 Free PMC article. Review.
References
-
- Adkins NL, Georgel PT. MeCP2: structure and functionThis paper is one of a selection of papers published in a Special Issue entitled 31st Annual International Asilomar Chromatin and Chromosomes Conference, and has undergone the Journal, As usual peer review process. Biochemistry and Cell Biology. 2011;89(1):1–11. - PubMed
-
- Amir RE, I, Van den Veyver B, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. - PubMed
-
- Bebbington A, Anderson A, Ravine D, Fyfe S, Pineda M, de Klerk N, Ben-Zeev B, Yatawara N, Percy A, Kaufmann WE, et al. Investigating genotype-phenotype relationships in Rett syndrome using an international data set. Neurology. 2008;70:868–875. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

