Insights from characterizing extinct human gut microbiomes
- PMID: 23251439
- PMCID: PMC3521025
- DOI: 10.1371/journal.pone.0051146
Insights from characterizing extinct human gut microbiomes
Abstract
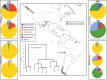
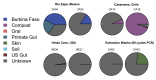
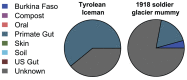
In an effort to better understand the ancestral state of the human distal gut microbiome, we examine feces retrieved from archaeological contexts (coprolites). To accomplish this, we pyrosequenced the 16S rDNA V3 region from duplicate coprolite samples recovered from three archaeological sites, each representing a different depositional environment: Hinds Cave (~8000 years B.P.) in the southern United States, Caserones (1600 years B.P.) in northern Chile, and Rio Zape in northern Mexico (1400 years B.P.). Clustering algorithms grouped samples from the same site. Phyletic representation was more similar within sites than between them. A Bayesian approach to source-tracking was used to compare the coprolite data to published data from known sources that include, soil, compost, human gut from rural African children, human gut, oral and skin from US cosmopolitan adults and non-human primate gut. The data from the Hinds Cave samples largely represented unknown sources. The Caserones samples, retrieved directly from natural mummies, matched compost in high proportion. A substantial and robust proportion of Rio Zape data was predicted to match the gut microbiome found in traditional rural communities, with more minor matches to other sources. One of the Rio Zape samples had taxonomic representation consistent with a child. To provide an idealized scenario for sample preservation, we also applied source tracking to previously published data for Ötzi the Iceman and a soldier frozen for 93 years on a glacier. Overall these studies reveal that human microbiome data has been preserved in some coprolites, and these preserved human microbiomes match more closely to those from the rural communities than to those from cosmopolitan communities. These results suggest that the modern cosmopolitan lifestyle resulted in a dramatic change to the human gut microbiome.
Conflict of interest statement
Figures

Similar articles
-
Taxonomic and predicted metabolic profiles of the human gut microbiome in pre-Columbian mummies.FEMS Microbiol Ecol. 2016 Nov;92(11):fiw182. doi: 10.1093/femsec/fiw182. Epub 2016 Aug 23. FEMS Microbiol Ecol. 2016. PMID: 27559027
-
Functional diversity of microbial ecologies estimated from ancient human coprolites and dental calculus.Philos Trans R Soc Lond B Biol Sci. 2020 Nov 23;375(1812):20190586. doi: 10.1098/rstb.2019.0586. Epub 2020 Oct 5. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 33012230 Free PMC article.
-
Human gut microbiome viewed across age and geography.Nature. 2012 May 9;486(7402):222-7. doi: 10.1038/nature11053. Nature. 2012. PMID: 22699611 Free PMC article.
-
Metagenomics of human microbiome: beyond 16s rDNA.Clin Microbiol Infect. 2012 Jul;18 Suppl 4:47-9. doi: 10.1111/j.1469-0691.2012.03865.x. Clin Microbiol Infect. 2012. PMID: 22647049 Review.
-
High throughput sequencing methods for microbiome profiling: application to food animal systems.Anim Health Res Rev. 2012 Jun;13(1):40-53. doi: 10.1017/S1466252312000126. Anim Health Res Rev. 2012. PMID: 22853944 Review.
Cited by
-
Microbial communities in pre-columbian coprolites.PLoS One. 2013 Jun 5;8(6):e65191. doi: 10.1371/journal.pone.0065191. Print 2013. PLoS One. 2013. PMID: 23755194 Free PMC article.
-
Ancient gut microbiomes shed light on modern disease.Environ Health Perspect. 2013 Apr;121(4):A118. doi: 10.1289/ehp.121-a118. Environ Health Perspect. 2013. PMID: 23548507 Free PMC article. No abstract available.
-
Estimating molecular preservation of the intestinal microbiome via metagenomic analyses of latrine sediments from two medieval cities.Philos Trans R Soc Lond B Biol Sci. 2020 Nov 23;375(1812):20190576. doi: 10.1098/rstb.2019.0576. Epub 2020 Oct 5. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 33012229 Free PMC article.
-
Understanding the gut microbiota by considering human evolution: a story of fire, cereals, cooking, molecular ingenuity, and functional cooperation.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0012722. doi: 10.1128/mmbr.00127-22. Epub 2023 Dec 21. Microbiol Mol Biol Rev. 2024. PMID: 38126754 Free PMC article. Review.
-
Biodiversity of protists and nematodes in the wild nonhuman primate gut.ISME J. 2020 Feb;14(2):609-622. doi: 10.1038/s41396-019-0551-4. Epub 2019 Nov 12. ISME J. 2020. PMID: 31719654 Free PMC article.
References
-
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI (2005) Host-bacterial mutualism in the human intestine. Science 307: 1915–1920. - PubMed
-
- Blaser M (2011) Antibiotic overuse: Stop the killing of beneficial bacteria. Nature 476: 393–394. - PubMed
-
- Neel JV, Weder AB, Julius S (1998) Type II diabetes, essential hypertension, and obesity as “syndromes of impaired genetic homeostasis”: the “thrifty genotype” hypothesis enters the 21st century. Perspectives in biology and medicine 42: 44–74. - PubMed

