Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design
- PMID: 23244564
- PMCID: PMC3646517
- DOI: 10.1021/jm301448p
Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design
Abstract
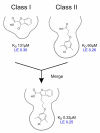
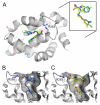
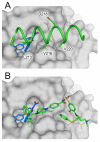
Myeloid cell leukemia 1 (Mcl-1), a member of the Bcl-2 family of proteins, is overexpressed and amplified in various cancers and promotes the aberrant survival of tumor cells that otherwise would undergo apoptosis. Here we describe the discovery of potent and selective Mcl-1 inhibitors using fragment-based methods and structure-based design. NMR-based screening of a large fragment library identified two chemically distinct hit series that bind to different sites on Mcl-1. Members of the two fragment classes were merged together to produce lead compounds that bind to Mcl-1 with a dissociation constant of <100 nM with selectivity for Mcl-1 over Bcl-xL and Bcl-2. Structures of merged compounds when complexed to Mcl-1 were obtained by X-ray crystallography and provide detailed information about the molecular recognition of small-molecule ligands binding Mcl-1. The compounds represent starting points for the discovery of clinically useful Mcl-1 inhibitors for the treatment of a wide variety of cancers.
Figures



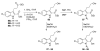

Comment in
-
Antagonists of protein-protein interactions made easy?J Med Chem. 2013 Jan 10;56(1):13-4. doi: 10.1021/jm301837n. Epub 2012 Dec 24. J Med Chem. 2013. PMID: 23265190 No abstract available.
Similar articles
-
Discovery of tricyclic indoles that potently inhibit Mcl-1 using fragment-based methods and structure-based design.J Med Chem. 2015 May 14;58(9):3794-805. doi: 10.1021/jm501984f. Epub 2015 Apr 17. J Med Chem. 2015. PMID: 25844895 Free PMC article.
-
Optimization of Potent and Selective Tricyclic Indole Diazepinone Myeloid Cell Leukemia-1 Inhibitors Using Structure-Based Design.J Med Chem. 2018 Mar 22;61(6):2410-2421. doi: 10.1021/acs.jmedchem.7b01155. Epub 2018 Mar 9. J Med Chem. 2018. PMID: 29323899
-
Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods.J Med Chem. 2016 Mar 10;59(5):2054-66. doi: 10.1021/acs.jmedchem.5b01660. Epub 2016 Feb 24. J Med Chem. 2016. PMID: 26878343 Free PMC article.
-
Recent Advances in Cancer Drug Development: Targeting Induced Myeloid Cell Leukemia-1 (Mcl-1) Differentiation Protein.Curr Med Chem. 2017;24(40):4488-4514. doi: 10.2174/0929867324666170912092659. Curr Med Chem. 2017. PMID: 28901269 Review.
-
Targeting cancer's Achilles' heel: role of BCL-2 inhibitors in cellular senescence and apoptosis.Future Med Chem. 2019 Sep;11(17):2287-2312. doi: 10.4155/fmc-2018-0366. Future Med Chem. 2019. PMID: 31581912 Review.
Cited by
-
Combined Physics- and Machine-Learning-Based Method to Identify Druggable Binding Sites Using SILCS-Hotspots.J Chem Inf Model. 2024 Oct 14;64(19):7743-7757. doi: 10.1021/acs.jcim.4c01189. Epub 2024 Sep 16. J Chem Inf Model. 2024. PMID: 39283165
-
Photodynamic Therapy with Tumor Cell Discrimination through RNA-Targeting Ability of Photosensitizer.Molecules. 2021 Oct 2;26(19):5990. doi: 10.3390/molecules26195990. Molecules. 2021. PMID: 34641533 Free PMC article.
-
Benzothiophene carboxylate derivatives as novel allosteric inhibitors of branched-chain α-ketoacid dehydrogenase kinase.J Biol Chem. 2014 Jul 25;289(30):20583-93. doi: 10.1074/jbc.M114.569251. J Biol Chem. 2014. PMID: 24895126 Free PMC article.
-
BM-1197: a novel and specific Bcl-2/Bcl-xL inhibitor inducing complete and long-lasting tumor regression in vivo.PLoS One. 2014 Jun 5;9(6):e99404. doi: 10.1371/journal.pone.0099404. eCollection 2014. PLoS One. 2014. PMID: 24901320 Free PMC article.
-
RASPD+: Fast Protein-Ligand Binding Free Energy Prediction Using Simplified Physicochemical Features.Front Mol Biosci. 2020 Dec 17;7:601065. doi: 10.3389/fmolb.2020.601065. eCollection 2020. Front Mol Biosci. 2020. PMID: 33392260 Free PMC article.
References
-
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. - PubMed
-
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluck RM, Adams JM, Huang DCS. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. - PubMed
-
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho Y-J, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao M-S, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials

