SnIPRE: selection inference using a Poisson random effects model
- PMID: 23236270
- PMCID: PMC3516574
- DOI: 10.1371/journal.pcbi.1002806
SnIPRE: selection inference using a Poisson random effects model
Abstract
We present an approach for identifying genes under natural selection using polymorphism and divergence data from synonymous and non-synonymous sites within genes. A generalized linear mixed model is used to model the genome-wide variability among categories of mutations and estimate its functional consequence. We demonstrate how the model's estimated fixed and random effects can be used to identify genes under selection. The parameter estimates from our generalized linear model can be transformed to yield population genetic parameter estimates for quantities including the average selection coefficient for new mutations at a locus, the synonymous and non-synynomous mutation rates, and species divergence times. Furthermore, our approach incorporates stochastic variation due to the evolutionary process and can be fit using standard statistical software. The model is fit in both the empirical Bayes and Bayesian settings using the lme4 package in R, and Markov chain Monte Carlo methods in WinBUGS. Using simulated data we compare our method to existing approaches for detecting genes under selection: the McDonald-Kreitman test, and two versions of the Poisson random field based method MKprf. Overall, we find our method universally outperforms existing methods for detecting genes subject to selection using polymorphism and divergence data.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
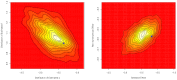
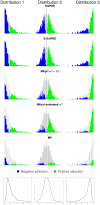
 used in the coalescent simulations for Table 7. Bottom: Proportion of constraint effects classified as significant by SnIPRE; x-axis is true proportion of non-lethal mutations,
used in the coalescent simulations for Table 7. Bottom: Proportion of constraint effects classified as significant by SnIPRE; x-axis is true proportion of non-lethal mutations,  .
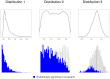
.
 .
.
 .
.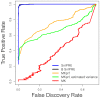
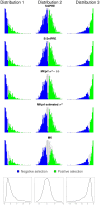
 or
or  .
.
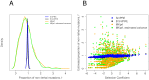
 ; the bottom row displays residuals for simulations using
; the bottom row displays residuals for simulations using  . Residuals grouped by true selection strength.
. Residuals grouped by true selection strength.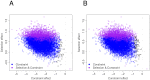
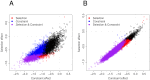
Similar articles
-
Statistical inference of selection and divergence from a time-dependent Poisson random field model.PLoS One. 2012;7(4):e34413. doi: 10.1371/journal.pone.0034413. Epub 2012 Apr 3. PLoS One. 2012. PMID: 22509300 Free PMC article.
-
Population genetics of polymorphism and divergence for diploid selection models with arbitrary dominance.Genetics. 2004 Sep;168(1):463-75. doi: 10.1534/genetics.103.024745. Genetics. 2004. PMID: 15454557 Free PMC article.
-
Identifying Lineage-Specific Targets of Natural Selection by a Bayesian Analysis of Genomic Polymorphisms and Divergence from Multiple Species.Mol Biol Evol. 2019 Jun 1;36(6):1302-1315. doi: 10.1093/molbev/msz046. Mol Biol Evol. 2019. PMID: 30840083
-
impMKT: the imputed McDonald and Kreitman test, a straightforward correction that significantly increases the evidence of positive selection of the McDonald and Kreitman test at the gene level.G3 (Bethesda). 2022 Sep 30;12(10):jkac206. doi: 10.1093/g3journal/jkac206. G3 (Bethesda). 2022. PMID: 35976111 Free PMC article. Review.
-
Applications of Monte Carlo Simulation in Modelling of Biochemical Processes.In: Mode CJ, editor. Applications of Monte Carlo Methods in Biology, Medicine and Other Fields of Science [Internet]. Rijeka (HR): InTech; 2011 Feb 28. Chapter 4. In: Mode CJ, editor. Applications of Monte Carlo Methods in Biology, Medicine and Other Fields of Science [Internet]. Rijeka (HR): InTech; 2011 Feb 28. Chapter 4. PMID: 28045483 Free Books & Documents. Review.
Cited by
-
Population genomic analysis reveals a rich speciation and demographic history of orang-utans (Pongo pygmaeus and Pongo abelii).PLoS One. 2013 Oct 23;8(10):e77175. doi: 10.1371/journal.pone.0077175. eCollection 2013. PLoS One. 2013. PMID: 24194868 Free PMC article.
-
Domestication in dry-cured meat Penicillium fungi: Convergent specific phenotypes and horizontal gene transfers without strong genetic subdivision.Evol Appl. 2023 Sep 8;16(9):1637-1660. doi: 10.1111/eva.13591. eCollection 2023 Sep. Evol Appl. 2023. PMID: 37752962 Free PMC article.
-
Genomic evidence for rediploidization and adaptive evolution following the whole-genome triplication.Nat Commun. 2024 Feb 22;15(1):1635. doi: 10.1038/s41467-024-46080-7. Nat Commun. 2024. PMID: 38388712 Free PMC article.
-
Pleiotropy constrains the evolution of protein but not regulatory sequences in a transcription regulatory network influencing complex social behaviors.Front Genet. 2014 Dec 23;5:431. doi: 10.3389/fgene.2014.00431. eCollection 2014. Front Genet. 2014. PMID: 25566318 Free PMC article.
-
Human OTULIN haploinsufficiency impairs cell-intrinsic immunity to staphylococcal α-toxin.Science. 2022 Jun 17;376(6599):eabm6380. doi: 10.1126/science.abm6380. Epub 2022 Jun 17. Science. 2022. PMID: 35587511 Free PMC article.
References
-
- Nielsen R (2005) Molecular signatures of natural selection. Genetics 39: 197. - PubMed
-
- Nielsen R (2001) Statistical tests of selective neutrality in the age of genomics. Heredity 86: 641–647. - PubMed
-
- McDonald J, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

