Endo-(1,4)-β-glucanase gene families in the grasses: temporal and spatial co-transcription of orthologous genes
- PMID: 23231659
- PMCID: PMC3557191
- DOI: 10.1186/1471-2229-12-235
Endo-(1,4)-β-glucanase gene families in the grasses: temporal and spatial co-transcription of orthologous genes
Abstract
Background: Endo-(1,4)-β-glucanase (cellulase) glycosyl hydrolase GH9 enzymes have been implicated in several aspects of cell wall metabolism in higher plants, including cellulose biosynthesis and degradation, modification of other wall polysaccharides that contain contiguous (1,4)-β-glucosyl residues, and wall loosening during cell elongation.
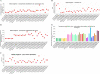
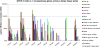
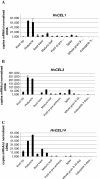
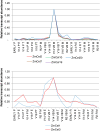
Results: The endo-(1,4)-β-glucanase gene families from barley (Hordeum vulgare), maize (Zea mays), sorghum (Sorghum bicolor), rice (Oryza sativa) and Brachypodium (Brachypodium distachyon) range in size from 23 to 29 members. Phylogenetic analyses show variations in clade structure between the grasses and Arabidopsis, and indicate differential gene loss and gain during evolution. Map positions and comparative studies of gene structures allow orthologous genes in the five species to be identified and synteny between the grasses is found to be high. It is also possible to differentiate between homoeologues resulting from ancient polyploidizations of the maize genome. Transcript analyses using microarray, massively parallel signature sequencing and quantitative PCR data for barley, rice and maize indicate that certain members of the endo-(1,4)-β-glucanase gene family are transcribed across a wide range of tissues, while others are specifically transcribed in particular tissues. There are strong correlations between transcript levels of several members of the endo-(1,4)-β-glucanase family and the data suggest that evolutionary conservation of transcription exists between orthologues across the grass family. There are also strong correlations between certain members of the endo-(1,4)-β-glucanase family and other genes known to be involved in cell wall loosening and cell expansion, such as expansins and xyloglucan endotransglycosylases.
Conclusions: The identification of these groups of genes will now allow us to test hypotheses regarding their functions and joint participation in wall synthesis, re-modelling and degradation, together with their potential role in lignocellulose conversion during biofuel production from grasses and cereal crop residues.
Figures


Similar articles
-
Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon.BMC Genomics. 2010 Oct 25;11:600. doi: 10.1186/1471-2164-11-600. BMC Genomics. 2010. PMID: 20973991 Free PMC article.
-
Review: The structure and function of cellulase (endo-β-1,4-glucanase) and hemicellulase (β-1,3-glucanase and endo-β-1,4-mannase) enzymes in invertebrates that consume materials ranging from microbes, algae to leaf litter.Comp Biochem Physiol B Biochem Mol Biol. 2020 Feb;240:110354. doi: 10.1016/j.cbpb.2019.110354. Epub 2019 Oct 21. Comp Biochem Physiol B Biochem Mol Biol. 2020. PMID: 31647988 Review.
-
Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution.Plant J. 2012 Aug;71(3):492-502. doi: 10.1111/j.1365-313X.2012.05005.x. Epub 2012 Jun 5. Plant J. 2012. PMID: 22443345
-
Heterologous expression and transcript analysis of gibberellin biosynthetic genes of grasses reveals novel functionality in the GA3ox family.BMC Plant Biol. 2015 Jun 5;15:130. doi: 10.1186/s12870-015-0520-7. BMC Plant Biol. 2015. PMID: 26044828 Free PMC article.
-
Multiple endo-1,4-beta-D-glucanase (cellulase) genes in Arabidopsis.Curr Top Dev Biol. 1999;46:39-61. doi: 10.1016/s0070-2153(08)60325-7. Curr Top Dev Biol. 1999. PMID: 10417876 Review.
Cited by
-
In silico Identification and Taxonomic Distribution of Plant Class C GH9 Endoglucanases.Front Plant Sci. 2016 Aug 12;7:1185. doi: 10.3389/fpls.2016.01185. eCollection 2016. Front Plant Sci. 2016. PMID: 27570528 Free PMC article.
-
Genome-wide analysis of the mulberry (Morus abla L.) GH9 gene family and the functional characterization of MaGH9B6 during the development of the abscission zone.Front Plant Sci. 2024 Apr 3;15:1352635. doi: 10.3389/fpls.2024.1352635. eCollection 2024. Front Plant Sci. 2024. PMID: 38633459 Free PMC article.
-
Action of an endo-β-1,3(4)-glucanase on cellobiosyl unit structure in barley β-1,3:1,4-glucan.Biosci Biotechnol Biochem. 2015;79(11):1810-7. doi: 10.1080/09168451.2015.1046365. Epub 2015 Jun 1. Biosci Biotechnol Biochem. 2015. PMID: 26027730 Free PMC article.
-
Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.Plant J. 2017 Feb;89(4):651-670. doi: 10.1111/tpj.13421. Epub 2017 Feb 11. Plant J. 2017. PMID: 27859885 Free PMC article.
-
A Genome-Wide Association Study for Culm Cellulose Content in Barley Reveals Candidate Genes Co-Expressed with Members of the CELLULOSE SYNTHASE A Gene Family.PLoS One. 2015 Jul 8;10(7):e0130890. doi: 10.1371/journal.pone.0130890. eCollection 2015. PLoS One. 2015. PMID: 26154104 Free PMC article.
References
-
- Burns JK, Lewandowski DJ, Nairn CJ, Brown GE. Endo 1,4-β-glucanase gene expression and cell wall hydrolase activities during abscission in Valencia orange. Physiol Plant. 1998;102(2):217–225. doi: 10.1034/j.1399-3054.1998.1020209.x. - DOI
-
- Ferrarese L, Moretto P, Trainotti L, Rascio N, Casadoro G. Cellulase involvement in the abscission of peach and pepper leaves is affected by salicylic acid. J Exp Bot. 1996;47(2):251–257. doi: 10.1093/jxb/47.2.251. - DOI
-
- Nunan KJ, Davies C, Robinson SP, Fincher GB. Expression patterns of cell wall-modifying enzymes during grape berry development. Planta. 2001;V214(2):257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

