Microarray-based gene expression profiling in patients with cryopyrin-associated periodic syndromes defines a disease-related signature and IL-1-responsive transcripts
- PMID: 23223423
- PMCID: PMC4174357
- DOI: 10.1136/annrheumdis-2012-202082
Microarray-based gene expression profiling in patients with cryopyrin-associated periodic syndromes defines a disease-related signature and IL-1-responsive transcripts
Abstract
Objective: To analyse gene expression patterns and to define a specific gene expression signature in patients with the severe end of the spectrum of cryopyrin-associated periodic syndromes (CAPS). The molecular consequences of interleukin 1 inhibition were examined by comparing gene expression patterns in 16 CAPS patients before and after treatment with anakinra.
Methods: We collected peripheral blood mononuclear cells from 22 CAPS patients with active disease and from 14 healthy children. Transcripts that passed stringent filtering criteria (p values≤false discovery rate 1%) were considered as differentially expressed genes (DEG). A set of DEG was validated by quantitative reverse transcription PCR and functional studies with primary cells from CAPS patients and healthy controls. We used 17 CAPS and 66 non-CAPS patient samples to create a set of gene expression models that differentiates CAPS patients from controls and from patients with other autoinflammatory conditions.
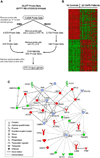
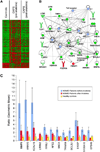
Results: Many DEG include transcripts related to the regulation of innate and adaptive immune responses, oxidative stress, cell death, cell adhesion and motility. A set of gene expression-based models comprising the CAPS-specific gene expression signature correctly classified all 17 samples from an independent dataset. This classifier also correctly identified 15 of 16 post-anakinra CAPS samples despite the fact that these CAPS patients were in clinical remission.
Conclusions: We identified a gene expression signature that clearly distinguished CAPS patients from controls. A number of DEG were in common with other systemic inflammatory diseases such as systemic onset juvenile idiopathic arthritis. The CAPS-specific gene expression classifiers also suggest incomplete suppression of inflammation at low doses of anakinra.
Keywords: Cytokines; Fever Syndromes; Inflammation.
Figures
Similar articles
-
Anakinra for cryopyrin-associated periodic syndrome.Expert Rev Clin Immunol. 2014 Jan;10(1):7-18. doi: 10.1586/1744666X.2014.861325. Epub 2013 Nov 21. Expert Rev Clin Immunol. 2014. PMID: 24308832 Review.
-
MRP8 and MRP14, phagocyte-specific danger signals, are sensitive biomarkers of disease activity in cryopyrin-associated periodic syndromes.Ann Rheum Dis. 2011 Dec;70(12):2075-2081. doi: 10.1136/ard.2011.152496. Epub 2011 Sep 8. Ann Rheum Dis. 2011. PMID: 21908452 Free PMC article.
-
Anakinra use during pregnancy in patients with cryopyrin-associated periodic syndromes (CAPS).Arthritis Rheumatol. 2014 Nov;66(11):3227-32. doi: 10.1002/art.38811. Arthritis Rheumatol. 2014. PMID: 25223501 Free PMC article.
-
Neonatal urticaria: Could it be CAPS?Pediatr Dermatol. 2018 Nov;35(6):e420-e421. doi: 10.1111/pde.13660. Epub 2018 Sep 6. Pediatr Dermatol. 2018. PMID: 30187963
-
Pharmacological treatment options for cryopyrin-associated periodic syndromes.Expert Rev Clin Pharmacol. 2017 Aug;10(8):855-864. doi: 10.1080/17512433.2017.1338946. Epub 2017 Jun 20. Expert Rev Clin Pharmacol. 2017. PMID: 28586272 Review.
Cited by
-
The Association of ATG16L1 Variations with Clinical Phenotypes of Adult-Onset Still's Disease.Genes (Basel). 2021 Jun 11;12(6):904. doi: 10.3390/genes12060904. Genes (Basel). 2021. PMID: 34208077 Free PMC article.
-
Gene Expression Analysis of Mevalonate Kinase Deficiency Affected Children Identifies Molecular Signatures Related to Hematopoiesis.Int J Environ Res Public Health. 2021 Jan 28;18(3):1170. doi: 10.3390/ijerph18031170. Int J Environ Res Public Health. 2021. PMID: 33525735 Free PMC article.
-
From bench to bedside and back again: translational research in autoinflammation.Nat Rev Rheumatol. 2015 Oct;11(10):573-85. doi: 10.1038/nrrheum.2015.79. Epub 2015 Jun 16. Nat Rev Rheumatol. 2015. PMID: 26077920 Review.
-
The Role of Epigenetics in Autoimmune/Inflammatory Disease.Front Immunol. 2019 Jul 4;10:1525. doi: 10.3389/fimmu.2019.01525. eCollection 2019. Front Immunol. 2019. PMID: 31333659 Free PMC article. Review.
-
Cell stress increases ATP release in NLRP3 inflammasome-mediated autoinflammatory diseases, resulting in cytokine imbalance.Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):2835-40. doi: 10.1073/pnas.1424741112. Epub 2015 Feb 17. Proc Natl Acad Sci U S A. 2015. PMID: 25730877 Free PMC article. Clinical Trial.
References
-
- Aksentijevich I, Kastner DL. Genetics of monogenic autoinflammatory diseases: past successes, future challenges. Nat Rev Rheumatol. 2011;7:469–478. - PubMed
-
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. - PubMed
-
- Agostini L, Martinon F, Burns K, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle–Wells autoinflammatory disorder. Immunity. 2004;20:319–325. - PubMed
-
- Tschopp J, Schroder K. NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
