High-resolution crystal structure of human protease-activated receptor 1
- PMID: 23222541
- PMCID: PMC3531875
- DOI: 10.1038/nature11701
High-resolution crystal structure of human protease-activated receptor 1
Abstract
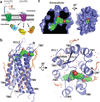
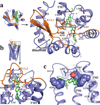
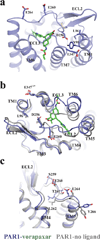
Protease-activated receptor 1 (PAR1) is the prototypical member of a family of G-protein-coupled receptors that mediate cellular responses to thrombin and related proteases. Thrombin irreversibly activates PAR1 by cleaving the amino-terminal exodomain of the receptor, which exposes a tethered peptide ligand that binds the heptahelical bundle of the receptor to affect G-protein activation. Here we report the 2.2 Å resolution crystal structure of human PAR1 bound to vorapaxar, a PAR1 antagonist. The structure reveals an unusual mode of drug binding that explains how a small molecule binds virtually irreversibly to inhibit receptor activation by the tethered ligand of PAR1. In contrast to deep, solvent-exposed binding pockets observed in other peptide-activated G-protein-coupled receptors, the vorapaxar-binding pocket is superficial but has little surface exposed to the aqueous solvent. Protease-activated receptors are important targets for drug development. The structure reported here will aid the development of improved PAR1 antagonists and the discovery of antagonists to other members of this receptor family.
Figures
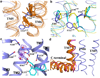
Similar articles
-
Entry from the Lipid Bilayer: A Possible Pathway for Inhibition of a Peptide G Protein-Coupled Receptor by a Lipophilic Small Molecule.Biochemistry. 2018 Oct 2;57(39):5748-5758. doi: 10.1021/acs.biochem.8b00577. Epub 2018 Aug 27. Biochemistry. 2018. PMID: 30102523 Free PMC article.
-
Investigating detailed interactions between novel PAR1 antagonist F16357 and the receptor using docking and molecular dynamic simulations.J Mol Graph Model. 2017 Oct;77:205-217. doi: 10.1016/j.jmgm.2017.08.019. Epub 2017 Aug 24. J Mol Graph Model. 2017. PMID: 28881236
-
Structural Properties of the Human Protease-Activated Receptor 1 Changing by a Strong Antagonist.Structure. 2018 Jun 5;26(6):829-838.e4. doi: 10.1016/j.str.2018.03.020. Epub 2018 May 3. Structure. 2018. PMID: 29731231
-
Targeting PAR1: Now What?Trends Pharmacol Sci. 2017 Aug;38(8):701-716. doi: 10.1016/j.tips.2017.05.001. Epub 2017 May 27. Trends Pharmacol Sci. 2017. PMID: 28558960 Free PMC article. Review.
-
From Multiple PAR1 Receptor/Protein Interactions to their Multiple Therapeutic Implications.Curr Top Med Chem. 2015;15(20):2080-114. doi: 10.2174/1568026615666150519103911. Curr Top Med Chem. 2015. PMID: 25986685 Review.
Cited by
-
G-protein-coupled receptors signaling pathways in new antiplatelet drug development.Arterioscler Thromb Vasc Biol. 2015 Mar;35(3):500-12. doi: 10.1161/ATVBAHA.114.303412. Epub 2015 Jan 29. Arterioscler Thromb Vasc Biol. 2015. PMID: 25633316 Free PMC article. Review.
-
Allosteric interactions in the parathyroid hormone GPCR-arrestin complex formation.Nat Chem Biol. 2020 Oct;16(10):1096-1104. doi: 10.1038/s41589-020-0567-0. Epub 2020 Jul 6. Nat Chem Biol. 2020. PMID: 32632293 Free PMC article.
-
Beyond standard molecular dynamics: investigating the molecular mechanisms of G protein-coupled receptors with enhanced molecular dynamics methods.Adv Exp Med Biol. 2014;796:95-125. doi: 10.1007/978-94-007-7423-0_6. Adv Exp Med Biol. 2014. PMID: 24158803 Free PMC article. Review.
-
Structure of the human P2Y12 receptor in complex with an antithrombotic drug.Nature. 2014 May 1;509(7498):115-8. doi: 10.1038/nature13083. Epub 2014 Mar 23. Nature. 2014. PMID: 24670650 Free PMC article.
-
Cofactoring and dimerization of proteinase-activated receptors.Pharmacol Rev. 2013 Sep 24;65(4):1198-213. doi: 10.1124/pr.111.004747. Print 2013. Pharmacol Rev. 2013. PMID: 24064459 Free PMC article. Review.
References
-
- Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. - PubMed
-
- Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. - PubMed
-
- Vu TK, Wheaton VI, Hung DT, Charo I, Coughlin SR. Domains specifying thrombin-receptor interaction. Nature. 1991;353:674–677. - PubMed
-
- Chen J, Ishii M, Wang L, Ishii K, Coughlin SR. Thrombin receptor activation. Confirmation of the intramolecular tethered liganding hypothesis and discovery of an alternative intermolecular liganding mode. J Biol Chem. 1994;269:16041–16045. - PubMed
-
- Liu LW, Vu TK, Esmon CT, Coughlin SR. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J Biol Chem. 1991;266:16977–16980. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

