Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals
- PMID: 23219535
- PMCID: PMC4151532
- DOI: 10.1016/j.molcel.2012.11.002
Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals
Abstract
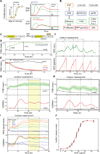
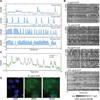
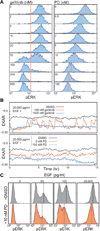
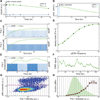
The EGF-stimulated ERK/MAPK pathway is a key conduit for cellular proliferation signals and a therapeutic target in many cancers. Here, we characterize two central quantitative aspects of this pathway: the mechanism by which signal strength is encoded and the response curve relating signal output to proliferation. Under steady-state conditions, we find that ERK is activated in discrete, asynchronous pulses with frequency and duration determined by extracellular concentrations of EGF spanning the physiological range. In genetically identical sister cells, cell-to-cell variability in pulse dynamics influences the decision to enter S phase. While targeted inhibition of EGFR reduces the frequency of ERK activity pulses, inhibition of MEK reduces their amplitude. Continuous response curves measured in multiple cell lines reveal that proliferation is effectively silenced only when ERK pathway output falls below a threshold of ~10%, indicating that high-dose targeting of the pathway is necessary to achieve therapeutic efficacy.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Differential effect of growth factors on invasion and proliferation of endocrine resistant breast cancer cells.PLoS One. 2012;7(7):e41847. doi: 10.1371/journal.pone.0041847. Epub 2012 Jul 30. PLoS One. 2012. PMID: 22860018 Free PMC article.
-
Structure of the EGF receptor transactivation circuit integrates multiple signals with cell context.Mol Biosyst. 2010 Jul;6(7):1293-306. doi: 10.1039/c003921g. Epub 2010 May 10. Mol Biosyst. 2010. PMID: 20458382 Free PMC article.
-
Role of Runx2 in crosstalk between Mek/Erk and PI3K/Akt signaling in MCF-10A cells.J Cell Biochem. 2014 Dec;115(12):2208-17. doi: 10.1002/jcb.24939. J Cell Biochem. 2014. PMID: 25147082
-
Blocking the PI3K/AKT and MEK/ERK signaling pathways can overcome gefitinib-resistance in non-small cell lung cancer cell lines.Adv Med Sci. 2011;56(2):275-84. doi: 10.2478/v10039-011-0043-x. Adv Med Sci. 2011. PMID: 22037177
-
Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer.Oncogene. 2007 May 14;26(22):3291-310. doi: 10.1038/sj.onc.1210422. Oncogene. 2007. PMID: 17496923 Review.
Cited by
-
Signaling, Deconstructed: Using Optogenetics to Dissect and Direct Information Flow in Biological Systems.Annu Rev Biomed Eng. 2021 Jul 13;23:61-87. doi: 10.1146/annurev-bioeng-083120-111648. Epub 2021 Mar 15. Annu Rev Biomed Eng. 2021. PMID: 33722063 Free PMC article. Review.
-
Enhanced kinase translocation reporters for simultaneous real-time measurement of PKA, ERK, and Ca2.bioRxiv [Preprint]. 2024 Oct 2:2024.09.30.615856. doi: 10.1101/2024.09.30.615856. bioRxiv. 2024. Update in: J Biol Chem. 2025 Jan 13:108183. doi: 10.1016/j.jbc.2025.108183 PMID: 39411162 Free PMC article. Updated. Preprint.
-
SALSA, a genetically encoded biosensor for spatiotemporal quantification of Notch signal transduction in vivo.Dev Cell. 2022 Apr 11;57(7):930-944.e6. doi: 10.1016/j.devcel.2022.03.008. Dev Cell. 2022. PMID: 35413239 Free PMC article.
-
Signalling dynamics, cell decisions, and homeostatic control in health and disease.Curr Opin Cell Biol. 2022 Apr;75:102066. doi: 10.1016/j.ceb.2022.01.011. Epub 2022 Mar 1. Curr Opin Cell Biol. 2022. PMID: 35245783 Free PMC article. Review.
-
The Fra-1/AP-1 Oncoprotein: From the "Undruggable" Transcription Factor to Therapeutic Targeting.Cancers (Basel). 2022 Mar 14;14(6):1480. doi: 10.3390/cancers14061480. Cancers (Basel). 2022. PMID: 35326630 Free PMC article. Review.
References
-
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. - PubMed
-
- Asthagiri AR, Reinhart CA, Horwitz AF, Lauffenburger DA. The role of transient ERK2 signals in fibronectin- and insulin-mediated DNA synthesis. J Cell Sci. 2000;113(Pt 24):4499–4510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

