Protocol: Chromatin immunoprecipitation (ChIP) methodology to investigate histone modifications in two model diatom species
- PMID: 23217141
- PMCID: PMC3546051
- DOI: 10.1186/1746-4811-8-48
Protocol: Chromatin immunoprecipitation (ChIP) methodology to investigate histone modifications in two model diatom species
Abstract
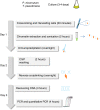
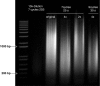
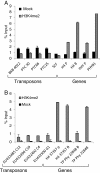
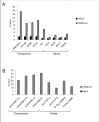
In this report we describe a chromatin immunoprecipitation (ChIP) protocol for two fully sequenced model diatom species Phaeodactylum tricornutum and Thalassiosira pseudonana. This protocol allows the extraction of satisfactory amounts of chromatin and gives reproducible results. We coupled the ChIP assay with real time quantitative PCR. Our results reveal that the two major histone marks H3K4me2 and H3K9me2 exist in P. tricornutum and T. pseudonana. As in other eukaryotes, H3K4me2 marks active genes whereas H3K9me2 marks transcriptionally inactive transposable elements. Unexpectedly however, T. pseudonana housekeeping genes also show a relative enrichment of H3K9me2. We also discuss optimization of the procedure, including growth conditions, cross linking and sonication. Validation of the protocol provides a set of genes and transposable elements that can be used as controls for studies using ChIP in each diatom species. This protocol can be easily adapted to other diatoms and eukaryotic phytoplankton species for genetic and biochemical studies.
Figures

Similar articles
-
Re-print of "Histone extraction protocol from the two model diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana".Mar Genomics. 2014 Aug;16:67-71. doi: 10.1016/j.margen.2014.05.003. Epub 2014 May 22. Mar Genomics. 2014. PMID: 24859489
-
Histone extraction protocol from the two model diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana.Mar Genomics. 2014 Feb;13:21-5. doi: 10.1016/j.margen.2013.11.006. Epub 2013 Dec 4. Mar Genomics. 2014. PMID: 24315927
-
An integrative analysis of post-translational histone modifications in the marine diatom Phaeodactylum tricornutum.Genome Biol. 2015 May 20;16(1):102. doi: 10.1186/s13059-015-0671-8. Genome Biol. 2015. PMID: 25990474 Free PMC article.
-
Genome editing in diatoms: achievements and goals.Plant Cell Rep. 2018 Oct;37(10):1401-1408. doi: 10.1007/s00299-018-2334-1. Epub 2018 Aug 23. Plant Cell Rep. 2018. PMID: 30167805 Review.
-
Oceanographic and biogeochemical insights from diatom genomes.Ann Rev Mar Sci. 2010;2:333-65. doi: 10.1146/annurev-marine-120308-081051. Ann Rev Mar Sci. 2010. PMID: 21141668 Review.
Cited by
-
Regulatory insight for a Zn2Cys6 transcription factor controlling effector-mediated virulence in a fungal pathogen of wheat.PLoS Pathog. 2024 Sep 23;20(9):e1012536. doi: 10.1371/journal.ppat.1012536. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39312592 Free PMC article.
-
Two Arabidopsis Homologs of Human Lysine-Specific Demethylase Function in Epigenetic Regulation of Plant Defense Responses.Front Plant Sci. 2021 Jun 14;12:688003. doi: 10.3389/fpls.2021.688003. eCollection 2021. Front Plant Sci. 2021. PMID: 34194459 Free PMC article.
-
Immunosuppressant MPA Modulates Tight Junction through Epigenetic Activation of MLCK/MLC-2 Pathway via p38MAPK.Front Physiol. 2015 Dec 22;6:381. doi: 10.3389/fphys.2015.00381. eCollection 2015. Front Physiol. 2015. Retraction in: Front Physiol. 2017 Sep 19;8:724. doi: 10.3389/fphys.2017.00724. PMID: 26733876 Free PMC article. Retracted.
-
An Efficient Chromatin Immunoprecipitation Protocol for the Analysis of Histone Modification Distributions in the Brown Alga Ectocarpus.Methods Protoc. 2022 Apr 25;5(3):36. doi: 10.3390/mps5030036. Methods Protoc. 2022. PMID: 35645344 Free PMC article.
-
Pathogen-Associated Molecular Pattern-Triggered Immunity Involves Proteolytic Degradation of Core Nonsense-Mediated mRNA Decay Factors During the Early Defense Response.Plant Cell. 2020 Apr;32(4):1081-1101. doi: 10.1105/tpc.19.00631. Epub 2020 Feb 21. Plant Cell. 2020. PMID: 32086363 Free PMC article.
References
-
- Round FE, Crawford RM, Mann DG. The diatoms: biology and morphology of the genera. London, UK: Cambridge University Press; 1990.
-
- Nelson DM, Treguer P, Brzezinski MA, Leynaert A, Queguiner B. Production and dissolution of biogenic silica in the ocean – revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycles. 1995;9:359–372. doi: 10.1029/95GB01070. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

