Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles
- PMID: 23209382
- PMCID: PMC3507959
- DOI: 10.1371/journal.pbio.1001442
Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles
Abstract
Interactions of cell-autonomous circadian oscillators with diurnal cycles govern the temporal compartmentalization of cell physiology in mammals. To understand the transcriptional and epigenetic basis of diurnal rhythms in mouse liver genome-wide, we generated temporal DNA occupancy profiles by RNA polymerase II (Pol II) as well as profiles of the histone modifications H3K4me3 and H3K36me3. We used these data to quantify the relationships of phases and amplitudes between different marks. We found that rhythmic Pol II recruitment at promoters rather than rhythmic transition from paused to productive elongation underlies diurnal gene transcription, a conclusion further supported by modeling. Moreover, Pol II occupancy preceded mRNA accumulation by 3 hours, consistent with mRNA half-lives. Both methylation marks showed that the epigenetic landscape is highly dynamic and globally remodeled during the 24-hour cycle. While promoters of transcribed genes had tri-methylated H3K4 even at their trough activity times, tri-methylation levels reached their peak, on average, 1 hour after Pol II. Meanwhile, rhythms in tri-methylation of H3K36 lagged transcription by 3 hours. Finally, modeling profiles of Pol II occupancy and mRNA accumulation identified three classes of genes: one showing rhythmicity both in transcriptional and mRNA accumulation, a second class with rhythmic transcription but flat mRNA levels, and a third with constant transcription but rhythmic mRNAs. The latter class emphasizes widespread temporally gated posttranscriptional regulation in the mouse liver.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
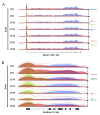
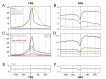
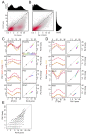

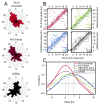
Comment in
-
Rhythmic changes in gene activation power the circadian clock.PLoS Biol. 2012;10(11):e1001443. doi: 10.1371/journal.pbio.1001443. Epub 2012 Nov 27. PLoS Biol. 2012. PMID: 23209383 Free PMC article. No abstract available.
Similar articles
-
Proteasome inhibition creates a chromatin landscape favorable to RNA Pol II processivity.J Biol Chem. 2020 Jan 31;295(5):1271-1287. doi: 10.1074/jbc.RA119.011174. Epub 2019 Dec 5. J Biol Chem. 2020. PMID: 31806706 Free PMC article.
-
Transcriptional regulatory logic of the diurnal cycle in the mouse liver.PLoS Biol. 2017 Apr 17;15(4):e2001069. doi: 10.1371/journal.pbio.2001069. eCollection 2017 Apr. PLoS Biol. 2017. PMID: 28414715 Free PMC article.
-
Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast.Genome Biol. 2011 Aug 22;12(8):R82. doi: 10.1186/gb-2011-12-8-r82. Genome Biol. 2011. PMID: 21859475 Free PMC article.
-
The RNA polymerase II transcriptional machinery and its epigenetic context.Subcell Biochem. 2013;61:237-59. doi: 10.1007/978-94-007-4525-4_11. Subcell Biochem. 2013. PMID: 23150254 Review.
-
How to stop: the mysterious links among RNA polymerase II occupancy 3' of genes, mRNA 3' processing and termination.Transcription. 2013 Jan-Feb;4(1):7-12. doi: 10.4161/trns.22300. Epub 2012 Nov 6. Transcription. 2013. PMID: 23131668 Free PMC article. Review.
Cited by
-
PRC2-EZH1 contributes to circadian gene expression by orchestrating chromatin states and RNA polymerase II complex stability.EMBO J. 2024 Oct 21. doi: 10.1038/s44318-024-00267-2. Online ahead of print. EMBO J. 2024. PMID: 39433902
-
Circadian regulation of translation.RNA Biol. 2024 Jan;21(1):14-24. doi: 10.1080/15476286.2024.2408524. Epub 2024 Sep 26. RNA Biol. 2024. PMID: 39324589 Free PMC article. Review.
-
Histone methylation: at the crossroad between circadian rhythms in transcription and metabolism.Front Genet. 2024 May 16;15:1343030. doi: 10.3389/fgene.2024.1343030. eCollection 2024. Front Genet. 2024. PMID: 38818037 Free PMC article. Review.
-
Diurnal control of iron responsive element containing mRNAs through iron regulatory proteins IRP1 and IRP2 is mediated by feeding rhythms.Genome Biol. 2024 May 21;25(1):128. doi: 10.1186/s13059-024-03270-2. Genome Biol. 2024. PMID: 38773499 Free PMC article.
-
Neuroinflammation and the role of epigenetic-based therapies for Huntington's disease management: the new paradigm.Inflammopharmacology. 2024 Jun;32(3):1791-1804. doi: 10.1007/s10787-024-01477-0. Epub 2024 Apr 23. Inflammopharmacology. 2024. PMID: 38653938 Review.
References
-
- Kornmann B, Schaad O, Bujard H, Takahashi J, Schibler U (2007) System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34 doi:10.1371/journal.pbio.0050034. - DOI - PMC - PubMed
-
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, et al. (2004) BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2: e377 doi: 10.1371/journal.pbio.0020377. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

