Computational model for autophagic vesicle dynamics in single cells
- PMID: 23196898
- PMCID: PMC3542220
- DOI: 10.4161/auto.22532
Computational model for autophagic vesicle dynamics in single cells
Abstract
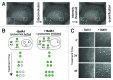
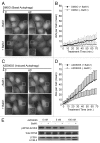
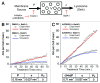
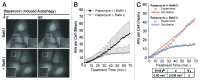
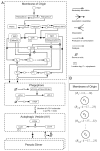
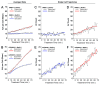
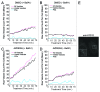
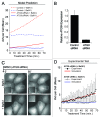
Macroautophagy (autophagy) is a cellular recycling program essential for homeostasis and survival during cytotoxic stress. This process, which has an emerging role in disease etiology and treatment, is executed in four stages through the coordinated action of more than 30 proteins. An effective strategy for studying complicated cellular processes, such as autophagy, involves the construction and analysis of mathematical or computational models. When developed and refined from experimental knowledge, these models can be used to interrogate signaling pathways, formulate novel hypotheses about systems, and make predictions about cell signaling changes induced by specific interventions. Here, we present the development of a computational model describing autophagic vesicle dynamics in a mammalian system. We used time-resolved, live-cell microscopy to measure the synthesis and turnover of autophagic vesicles in single cells. The stochastically simulated model was consistent with data acquired during conditions of both basal and chemically-induced autophagy. The model was tested by genetic modulation of autophagic machinery and found to accurately predict vesicle dynamics observed experimentally. Furthermore, the model generated an unforeseen prediction about vesicle size that is consistent with both published findings and our experimental observations. Taken together, this model is accurate and useful and can serve as the foundation for future efforts aimed at quantitative characterization of autophagy.
Figures
Similar articles
-
Agent-based modeling of autophagy reveals emergent regulatory behavior of spatio-temporal autophagy dynamics.Cell Commun Signal. 2014 Sep 10;12:56. doi: 10.1186/s12964-014-0056-8. Cell Commun Signal. 2014. PMID: 25214434 Free PMC article.
-
Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells.Traffic. 2008 Feb;9(2):230-50. doi: 10.1111/j.1600-0854.2007.00677.x. Epub 2007 Dec 7. Traffic. 2008. PMID: 17999726
-
Cannabinoid receptor 1 modulates the autophagic flux independent of mTOR- and BECLIN1-complex.J Neurochem. 2014 Nov;131(4):484-97. doi: 10.1111/jnc.12839. Epub 2014 Aug 26. J Neurochem. 2014. PMID: 25066892
-
Molecules and their functions in autophagy.Exp Mol Med. 2012 Feb 29;44(2):73-80. doi: 10.3858/emm.2012.44.2.029. Exp Mol Med. 2012. PMID: 22257882 Free PMC article. Review.
-
Trafficking and signaling in mammalian autophagy.IUBMB Life. 2010 Jul;62(7):503-8. doi: 10.1002/iub.334. IUBMB Life. 2010. PMID: 20552641 Review.
Cited by
-
PI3K-C2α knockdown decreases autophagy and maturation of endocytic vesicles.PLoS One. 2017 Sep 14;12(9):e0184909. doi: 10.1371/journal.pone.0184909. eCollection 2017. PLoS One. 2017. PMID: 28910396 Free PMC article.
-
Modeling of autophagy-related gene expression dynamics during long term fasting in European eel (Anguilla anguilla).Sci Rep. 2017 Dec 20;7(1):17896. doi: 10.1038/s41598-017-18164-6. Sci Rep. 2017. PMID: 29263413 Free PMC article.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.Autophagy. 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. Autophagy. 2021. PMID: 33634751 Free PMC article.
-
A Potent and Selective ULK1 Inhibitor Suppresses Autophagy and Sensitizes Cancer Cells to Nutrient Stress.iScience. 2018 Oct 26;8:74-84. doi: 10.1016/j.isci.2018.09.012. Epub 2018 Sep 19. iScience. 2018. PMID: 30292171 Free PMC article.
-
Agent-based modeling of autophagy reveals emergent regulatory behavior of spatio-temporal autophagy dynamics.Cell Commun Signal. 2014 Sep 10;12:56. doi: 10.1186/s12964-014-0056-8. Cell Commun Signal. 2014. PMID: 25214434 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
