Identification and profiling of CXCR3-CXCR4 chemokine receptor heteromer complexes
- PMID: 23170857
- PMCID: PMC3605874
- DOI: 10.1111/bph.12064
Identification and profiling of CXCR3-CXCR4 chemokine receptor heteromer complexes
Abstract
Background and purpose: The C-X-C chemokine receptors 3 (CXCR3) and C-X-C chemokine receptors 4 (CXCR4) are involved in various autoimmune diseases and cancers. Small antagonists have previously been shown to cross-inhibit chemokine binding to CXCR4, CC chemokine receptors 2 (CCR2) and 5 (CCR5) heteromers. We investigated whether CXCR3 and CXCR4 can form heteromeric complexes and the binding characteristics of chemokines and small ligand compounds to these chemokine receptor heteromers.
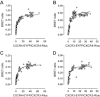
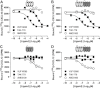
Experimental approach: CXCR3-CXCR4 heteromers were identified in HEK293T cells using co-immunoprecipitation, time-resolved fluorescence resonance energy transfer, saturation BRET and the GPCR-heteromer identification technology (HIT) approach. Equilibrium competition binding and dissociation experiments were performed to detect negative binding cooperativity.
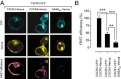
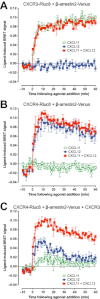
Key results: We provide evidence that chemokine receptors CXCR3 and CXCR4 form heteromeric complexes in HEK293T cells. Chemokine binding was mutually exclusive on membranes co-expressing CXCR3 and CXCR4 as revealed by equilibrium competition binding and dissociation experiments. The small CXCR3 agonist VUF10661 impaired binding of CXCL12 to CXCR4, whereas small antagonists were unable to cross-inhibit chemokine binding to the other chemokine receptor. In contrast, negative binding cooperativity between CXCR3 and CXCR4 chemokines was not observed in intact cells. However, using the GPCR-HIT approach, we have evidence for specific β-arrestin2 recruitment to CXCR3-CXCR4 heteromers in response to agonist stimulation.
Conclusions and implications: This study indicates that heteromeric CXCR3-CXCR4 complexes may act as functional units in living cells, which potentially open up novel therapeutic opportunities.
© 2012 The Authors. British Journal of Pharmacology © 2012 The British Pharmacological Society.
Figures



Similar articles
-
Pharmacological characterization of a small-molecule agonist for the chemokine receptor CXCR3.Br J Pharmacol. 2012 Jun;166(3):898-911. doi: 10.1111/j.1476-5381.2011.01648.x. Br J Pharmacol. 2012. PMID: 21883151 Free PMC article.
-
Characterization of heteromeric complexes between chemokine (C-X-C motif) receptor 4 and α1-adrenergic receptors utilizing intermolecular bioluminescence resonance energy transfer assays.Biochem Biophys Res Commun. 2020 Jul 23;528(2):368-375. doi: 10.1016/j.bbrc.2020.02.094. Epub 2020 Feb 19. Biochem Biophys Res Commun. 2020. PMID: 32085899 Free PMC article.
-
Asymmetrical ligand-induced cross-regulation of chemokine (C-X-C motif) receptor 4 by α1-adrenergic receptors at the heteromeric receptor complex.Sci Rep. 2018 Feb 9;8(1):2730. doi: 10.1038/s41598-018-21096-4. Sci Rep. 2018. PMID: 29426850 Free PMC article.
-
Chemokine receptor trio: CXCR3, CXCR4 and CXCR7 crosstalk via CXCL11 and CXCL12.Cytokine Growth Factor Rev. 2013 Feb;24(1):41-9. doi: 10.1016/j.cytogfr.2012.08.007. Epub 2012 Sep 16. Cytokine Growth Factor Rev. 2013. PMID: 22989616 Free PMC article. Review.
-
Small Molecule CXCR3 Antagonists.J Med Chem. 2016 Apr 14;59(7):2894-917. doi: 10.1021/acs.jmedchem.5b01337. Epub 2015 Nov 4. J Med Chem. 2016. PMID: 26535614 Review.
Cited by
-
Chemokinergic and Dopaminergic Signalling Collaborates through the Heteromer Formed by CCR9 and Dopamine Receptor D5 Increasing the Migratory Speed of Effector CD4+ T-Cells to Infiltrate the Colonic Mucosa.Int J Mol Sci. 2024 Sep 18;25(18):10022. doi: 10.3390/ijms251810022. Int J Mol Sci. 2024. PMID: 39337509 Free PMC article.
-
Profiling epidermal growth factor receptor and heregulin receptor 3 heteromerization using receptor tyrosine kinase heteromer investigation technology.PLoS One. 2013 May 20;8(5):e64672. doi: 10.1371/journal.pone.0064672. Print 2013. PLoS One. 2013. PMID: 23700486 Free PMC article.
-
Coexpression of CCR7 and CXCR4 During B Cell Development Controls CXCR4 Responsiveness and Bone Marrow Homing.Front Immunol. 2019 Dec 18;10:2970. doi: 10.3389/fimmu.2019.02970. eCollection 2019. Front Immunol. 2019. PMID: 31921208 Free PMC article.
-
New Insights into Mechanisms and Functions of Chemokine (C-X-C Motif) Receptor 4 Heteromerization in Vascular Smooth Muscle.Int J Mol Sci. 2016 Jun 20;17(5):971. doi: 10.3390/ijms17060971. Int J Mol Sci. 2016. PMID: 27331810 Free PMC article.
-
Functional interaction between angiotensin II receptor type 1 and chemokine (C-C motif) receptor 2 with implications for chronic kidney disease.PLoS One. 2015 Mar 25;10(3):e0119803. doi: 10.1371/journal.pone.0119803. eCollection 2015. PLoS One. 2015. PMID: 25807547 Free PMC article.
References
-
- Babcock GJ, Farzan M, Sodroski J. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J Biol Chem. 2003;278:3378–3385. - PubMed
-
- Birdsall NJ. Class A GPCR heterodimers: evidence from binding studies. Trends Pharmacol Sci. 2010;31:499–508. - PubMed
-
- Chabre M, Deterre P, Antonny B. The apparent cooperativity of some GPCRs does not necessarily imply dimerization. Trends Pharmacol Sci. 2009;30:182–187. - PubMed
-
- Christopoulos A, Lanzafame A, Ziegler A, Mitchelson F. Kinetic studies of co-operativity at atrial muscarinic M2 receptors with an ‘infinite dilution’ procedure. Biochem Pharmacol. 1997;53:795–800. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

