Tet1 controls meiosis by regulating meiotic gene expression
- PMID: 23151479
- PMCID: PMC3528851
- DOI: 10.1038/nature11709
Tet1 controls meiosis by regulating meiotic gene expression
Abstract
Meiosis is a germ-cell-specific cell division process through which haploid gametes are produced for sexual reproduction. Before the initiation of meiosis, mouse primordial germ cells undergo a series of epigenetic reprogramming steps, including the global erasure of DNA methylation at the 5-position of cytosine (5mC) in CpG-rich DNA. Although several epigenetic regulators, such as Dnmt3l and the histone methyltransferases G9a and Prdm9, have been reported to be crucial for meiosis, little is known about how the expression of meiotic genes is regulated and how their expression contributes to normal meiosis. Using a loss-of-function approach in mice, here we show that the 5mC-specific dioxygenase Tet1 has an important role in regulating meiosis in mouse oocytes. Tet1 deficiency significantly reduces female germ-cell numbers and fertility. Univalent chromosomes and unresolved DNA double-strand breaks are also observed in Tet1-deficient oocytes. Tet1 deficiency does not greatly affect the genome-wide demethylation that takes place in primordial germ cells, but leads to defective DNA demethylation and decreased expression of a subset of meiotic genes. Our study thus establishes a function for Tet1 in meiosis and meiotic gene activation in female germ cells.
Figures
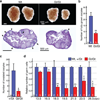
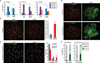
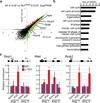
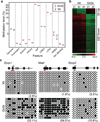
Comment in
-
Epigenetics: Erase for a new start.Nature. 2012 Dec 20;492(7429):363-4. doi: 10.1038/492363a. Nature. 2012. PMID: 23257876 No abstract available.
Similar articles
-
Role of Tet1 in erasure of genomic imprinting.Nature. 2013 Dec 19;504(7480):460-4. doi: 10.1038/nature12805. Epub 2013 Dec 1. Nature. 2013. PMID: 24291790 Free PMC article.
-
The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes.Nature. 2011 Sep 4;477(7366):606-10. doi: 10.1038/nature10443. Nature. 2011. PMID: 21892189
-
Iterative oxidation by TET1 is required for reprogramming of imprinting control regions and patterning of mouse sperm hypomethylated regions.Dev Cell. 2024 Apr 22;59(8):1010-1027.e8. doi: 10.1016/j.devcel.2024.02.012. Epub 2024 Apr 2. Dev Cell. 2024. PMID: 38569549
-
The Regulation and Function of Cohesin and Condensin in Mammalian Oocytes and Spermatocytes.Results Probl Cell Differ. 2017;63:355-372. doi: 10.1007/978-3-319-60855-6_15. Results Probl Cell Differ. 2017. PMID: 28779325 Review.
-
The Beginning of Meiosis in Mammalian Female Germ Cells: A Never-Ending Story of Intrinsic and Extrinsic Factors.Int J Mol Sci. 2022 Oct 20;23(20):12571. doi: 10.3390/ijms232012571. Int J Mol Sci. 2022. PMID: 36293427 Free PMC article. Review.
Cited by
-
TET1 modulates H4K16 acetylation by controlling auto-acetylation of hMOF to affect gene regulation and DNA repair function.Nucleic Acids Res. 2017 Jan 25;45(2):672-684. doi: 10.1093/nar/gkw919. Epub 2016 Oct 12. Nucleic Acids Res. 2017. PMID: 27733505 Free PMC article.
-
CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases.Mol Cell. 2015 Jan 22;57(2):247-260. doi: 10.1016/j.molcel.2014.12.002. Epub 2014 Dec 31. Mol Cell. 2015. PMID: 25557551 Free PMC article.
-
DNA Methylation Reprogramming during Mammalian Development.Genes (Basel). 2019 Mar 29;10(4):257. doi: 10.3390/genes10040257. Genes (Basel). 2019. PMID: 30934924 Free PMC article. Review.
-
Ascorbic acid induces global epigenetic reprogramming to promote meiotic maturation and developmental competence of porcine oocytes.Sci Rep. 2018 Apr 17;8(1):6132. doi: 10.1038/s41598-018-24395-y. Sci Rep. 2018. PMID: 29666467 Free PMC article.
-
Mechanism and function of oxidative reversal of DNA and RNA methylation.Annu Rev Biochem. 2014;83:585-614. doi: 10.1146/annurev-biochem-060713-035513. Annu Rev Biochem. 2014. PMID: 24905787 Free PMC article. Review.
References
-
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nature Reviews Genetics. 2010;11:124–136. - PubMed
-
- Hayashi K, Surani MA. Resetting the epigenome beyond pluripotency in the germline. Cell Stem Cell. 2009;4:493–498. - PubMed
-
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;2008:129–140. - PubMed
-
- Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. - PubMed
-
- Seki Y, et al. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. - PubMed
References for Methods
-
- Markoulaki S, Meissner A, Jaenisch R. Somatic cell nuclear transfer and derivation of embryonic stem cells in the mouse. Methods (San Diego, Calif.) 2008;45:101–114. - PubMed
-
- Kouznetsova A, Novak I, Jessberger R, Hoog C. SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J Cell Sci. 2005;118:2271–2278. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

