Rev1 recruits ung to switch regions and enhances du glycosylation for immunoglobulin class switch DNA recombination
- PMID: 23140944
- PMCID: PMC3518390
- DOI: 10.1016/j.celrep.2012.09.029
Rev1 recruits ung to switch regions and enhances du glycosylation for immunoglobulin class switch DNA recombination
Abstract
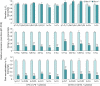
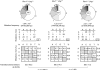
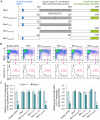
By diversifying the biological effector functions of antibodies, class switch DNA recombination (CSR) plays a critical role in the maturation of the immune response. It is initiated by activation-induced cytidine deaminase (AID)-mediated deoxycytosine deamination, yielding deoxyuridine (dU), and dU glycosylation by uracil DNA glycosylase (Ung) in antibody switch (S) region DNA. Here we showed that the translesion DNA synthesis polymerase Rev1 directly interacted with Ung and targeted in an AID-dependent and Ung-independent fashion the S regions undergoing CSR. Rev1(-/-)Ung(+/+) B cells reduced Ung recruitment to S regions, DNA-dU glycosylation, and CSR. Together with an S region spectrum of mutations similar to that of Rev1(+/+)Ung(-/-) B cells, this suggests that Rev1 operates in the same pathway as Ung, as emphasized by further decreased CSR in Rev1(-/-)Msh2(-/-) B cells. Rescue of CSR in Rev1(-/-) B cells by a catalytically inactive Rev1 mutant shows that the important role of Rev1 in CSR is mediated by Rev1's scaffolding function, not its enzymatic function.
Copyright © 2012 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
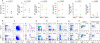



Similar articles
-
AID- and Ung-dependent generation of staggered double-strand DNA breaks in immunoglobulin class switch DNA recombination: a post-cleavage role for AID.Mol Immunol. 2008 Nov;46(1):45-61. doi: 10.1016/j.molimm.2008.07.003. Epub 2008 Aug 28. Mol Immunol. 2008. PMID: 18760480 Free PMC article.
-
Rev1 is essential in generating G to C transversions downstream of the Ung2 pathway but not the Msh2+Ung2 hybrid pathway.Eur J Immunol. 2013 Oct;43(10):2765-70. doi: 10.1002/eji.201243191. Epub 2013 Aug 5. Eur J Immunol. 2013. PMID: 23857323
-
Single-strand DNA breaks in Ig class switch recombination that depend on UNG but not AID.Int Immunol. 2008 Nov;20(11):1381-93. doi: 10.1093/intimm/dxn097. Epub 2008 Sep 15. Int Immunol. 2008. PMID: 18794203
-
Opinion: uracil DNA glycosylase (UNG) plays distinct and non-canonical roles in somatic hypermutation and class switch recombination.Int Immunol. 2014 Oct;26(10):575-8. doi: 10.1093/intimm/dxu071. Epub 2014 Jul 3. Int Immunol. 2014. PMID: 24994819 Free PMC article. Review.
-
Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies.Adv Immunol. 2007;94:275-306. doi: 10.1016/S0065-2776(06)94009-7. Adv Immunol. 2007. PMID: 17560278 Review.
Cited by
-
Epigenetics of Peripheral B-Cell Differentiation and the Antibody Response.Front Immunol. 2015 Dec 14;6:631. doi: 10.3389/fimmu.2015.00631. eCollection 2015. Front Immunol. 2015. PMID: 26697022 Free PMC article. Review.
-
Generating and repairing genetically programmed DNA breaks during immunoglobulin class switch recombination.F1000Res. 2018 Apr 13;7:458. doi: 10.12688/f1000research.13247.1. eCollection 2018. F1000Res. 2018. PMID: 29744038 Free PMC article. Review.
-
Histone deacetylase inhibitors upregulate B cell microRNAs that silence AID and Blimp-1 expression for epigenetic modulation of antibody and autoantibody responses.J Immunol. 2014 Dec 15;193(12):5933-50. doi: 10.4049/jimmunol.1401702. Epub 2014 Nov 12. J Immunol. 2014. PMID: 25392531 Free PMC article.
-
Activation-induced Cytidine Deaminase in B Cell Immunity and Cancers.Immune Netw. 2012 Dec;12(6):230-9. doi: 10.4110/in.2012.12.6.230. Epub 2012 Dec 31. Immune Netw. 2012. PMID: 23396757 Free PMC article.
-
Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes.Proc Natl Acad Sci U S A. 2013 Jul 2;110(27):E2470-9. doi: 10.1073/pnas.1308512110. Epub 2013 Jun 10. Proc Natl Acad Sci U S A. 2013. PMID: 23754438 Free PMC article.
References
-
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

