Impact of copy number variations (CNVs) on long-range gene regulation at the HoxD locus
- PMID: 23134724
- PMCID: PMC3528568
- DOI: 10.1073/pnas.1217659109
Impact of copy number variations (CNVs) on long-range gene regulation at the HoxD locus
Abstract
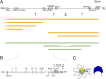
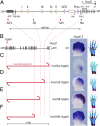
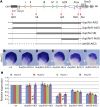
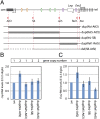
Copy number variations are genomic structural variants that are frequently associated with human diseases. Among these copy number variations, duplications of DNA segments are often assumed to lead to dosage effects by increasing the copy number of either genes or their regulatory elements. We produced a series of large targeted duplications within a conserved gene desert upstream of the murine HoxD locus. This DNA region, syntenic to human 2q31-32, contains a range of regulatory elements required for Hoxd gene transcription, and it is often disrupted and/or reorganized in human genetic conditions collectively known as the 2q31 syndrome. Unexpectedly, one such duplication led to a transcriptional down-regulation in developing digits by impairing physical interactions between the target genes and their upstream regulatory elements, thus phenocopying the effect obtained when these enhancer sequences are deleted. These results illustrate the detrimental consequences of interrupting highly conserved regulatory landscapes and reveal a mechanism where genomic duplications lead to partial loss of function of nearby located genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs.Nature. 2002 Nov 14;420(6912):145-50. doi: 10.1038/nature01189. Nature. 2002. PMID: 12432383
-
An enhancer-titration effect induces digit-specific regulatory alleles of the HoxD cluster.Dev Biol. 2003 Apr 15;256(2):212-20. doi: 10.1016/s0012-1606(02)00136-7. Dev Biol. 2003. PMID: 12679098
-
Targeted inversion of a polar silencer within the HoxD complex re-allocates domains of enhancer sharing.Nat Genet. 2000 Dec;26(4):451-4. doi: 10.1038/82593. Nat Genet. 2000. PMID: 11101844
-
Copy-number variations, noncoding sequences, and human phenotypes.Annu Rev Genomics Hum Genet. 2011;12:53-72. doi: 10.1146/annurev-genom-082410-101404. Annu Rev Genomics Hum Genet. 2011. PMID: 21756107 Review.
-
An Evolutionary Perspective on the Impact of Genomic Copy Number Variation on Human Health.J Mol Evol. 2020 Jan;88(1):104-119. doi: 10.1007/s00239-019-09911-6. Epub 2019 Sep 14. J Mol Evol. 2020. PMID: 31522275 Review.
Cited by
-
Evolutionary stability of topologically associating domains is associated with conserved gene regulation.BMC Biol. 2018 Aug 7;16(1):87. doi: 10.1186/s12915-018-0556-x. BMC Biol. 2018. PMID: 30086749 Free PMC article.
-
Regulation of disease-associated gene expression in the 3D genome.Nat Rev Mol Cell Biol. 2016 Dec;17(12):771-782. doi: 10.1038/nrm.2016.138. Epub 2016 Nov 9. Nat Rev Mol Cell Biol. 2016. PMID: 27826147 Review.
-
Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding.Genome Res. 2017 Feb;27(2):223-233. doi: 10.1101/gr.213066.116. Epub 2016 Dec 6. Genome Res. 2017. PMID: 27923844 Free PMC article.
-
A discrete transition zone organizes the topological and regulatory autonomy of the adjacent tfap2c and bmp7 genes.PLoS Genet. 2015 Jan 8;11(1):e1004897. doi: 10.1371/journal.pgen.1004897. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569170 Free PMC article.
-
Three-dimensional genome structure and function.MedComm (2020). 2023 Jul 8;4(4):e326. doi: 10.1002/mco2.326. eCollection 2023 Aug. MedComm (2020). 2023. PMID: 37426677 Free PMC article. Review.
References
-
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. - PubMed
-
- Klopocki E, Mundlos S. Copy-number variations, noncoding sequences, and human phenotypes. Annu Rev Genomics Hum Genet. 2011;12:53–72. - PubMed
-
- Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. - PubMed
-
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132(4):797–803. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

