Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation
- PMID: 23115248
- PMCID: PMC3517233
- DOI: 10.1105/tpc.112.103838
Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation
Erratum in
- Plant Cell. 2012 Dec;24(12):5193
Abstract
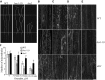
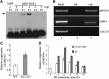
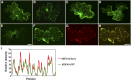
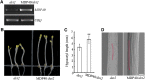
The brassinosteroid (BR) phytohormones play crucial roles in regulating plant cell growth and morphogenesis, particularly in hypocotyl cell elongation. The microtubule cytoskeleton is also known to participate in the regulation of hypocotyl elongation. However, it is unclear if BR regulation of hypocotyl elongation involves the microtubule cytoskeleton. In this study, we demonstrate that BRs mediate hypocotyl cell elongation by influencing the orientation and stability of cortical microtubules. Further analysis identified the previously undiscovered Arabidopsis thaliana microtubule destabilizing protein40 (MDP40) as a positive regulator of hypocotyl cell elongation. Brassinazole-resistant1, a key transcription factor in the BR signaling pathway, directly targets and upregulates MDP40. Overexpression of MDP40 partially rescued the shorter hypocotyl phenotype in BR-deficient mutant de-etiolated-2 seedlings. Reorientation of the cortical microtubules in the cells of MDP40 RNA interference transgenic lines was less sensitive to BR. These findings demonstrate that MDP40 is a key regulator in BR regulation of cortical microtubule reorientation and mediates hypocotyl growth. This study reveals a mechanism involving BR regulation of microtubules through MDP40 to mediate hypocotyl cell elongation.
Figures


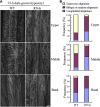
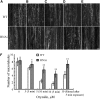

Similar articles
-
Ethylene Regulates the Arabidopsis Microtubule-Associated Protein WAVE-DAMPENED2-LIKE5 in Etiolated Hypocotyl Elongation.Plant Physiol. 2015 Sep;169(1):325-37. doi: 10.1104/pp.15.00609. Epub 2015 Jul 1. Plant Physiol. 2015. PMID: 26134166 Free PMC article.
-
MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis.Plant Cell. 2011 Dec;23(12):4411-27. doi: 10.1105/tpc.111.092684. Epub 2011 Dec 30. Plant Cell. 2011. PMID: 22209764 Free PMC article.
-
COP1 mediates dark-specific degradation of microtubule-associated protein WDL3 in regulating Arabidopsis hypocotyl elongation.Proc Natl Acad Sci U S A. 2017 Nov 14;114(46):12321-12326. doi: 10.1073/pnas.1708087114. Epub 2017 Oct 30. Proc Natl Acad Sci U S A. 2017. PMID: 29087315 Free PMC article.
-
Coordinated Regulation of Hypocotyl Cell Elongation by Light and Ethylene through a Microtubule Destabilizing Protein.Plant Physiol. 2018 Jan;176(1):678-690. doi: 10.1104/pp.17.01109. Epub 2017 Nov 22. Plant Physiol. 2018. PMID: 29167353 Free PMC article.
-
Cytoplasmic Linker Protein-Associating Protein at the Nexus of Hormone Signaling, Microtubule Organization, and the Transition From Division to Differentiation in Primary Roots.Front Plant Sci. 2022 Apr 28;13:883363. doi: 10.3389/fpls.2022.883363. eCollection 2022. Front Plant Sci. 2022. PMID: 35574108 Free PMC article. Review.
Cited by
-
The microtubule-associated protein WDL4 modulates auxin distribution to promote apical hook opening in Arabidopsis.Plant Cell. 2021 Jul 19;33(6):1927-1944. doi: 10.1093/plcell/koab080. Plant Cell. 2021. PMID: 33730147 Free PMC article.
-
Ethylene Regulates the Arabidopsis Microtubule-Associated Protein WAVE-DAMPENED2-LIKE5 in Etiolated Hypocotyl Elongation.Plant Physiol. 2015 Sep;169(1):325-37. doi: 10.1104/pp.15.00609. Epub 2015 Jul 1. Plant Physiol. 2015. PMID: 26134166 Free PMC article.
-
Cortical microtubule rearrangements and cell wall patterning.Front Plant Sci. 2015 Apr 8;6:236. doi: 10.3389/fpls.2015.00236. eCollection 2015. Front Plant Sci. 2015. PMID: 25904930 Free PMC article.
-
HY5 inhibits lateral root initiation in Arabidopsis through negative regulation of the microtubule-stabilizing protein TPXL5.Plant Cell. 2023 Mar 15;35(3):1092-1109. doi: 10.1093/plcell/koac358. Plant Cell. 2023. PMID: 36512471 Free PMC article.
-
Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell wall patterns.Plant Cell. 2013 Nov;25(11):4439-50. doi: 10.1105/tpc.113.117853. Epub 2013 Nov 26. Plant Cell. 2013. PMID: 24280391 Free PMC article.
References
-
- Buschmann H., Lloyd C.W. (2008). Arabidopsis mutants and the network of microtubule-associated functions. Mol. Plant 1: 888–898 - PubMed
-
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

