Alternative transcription start site selection leads to large differences in translation activity in yeast
- PMID: 23105001
- PMCID: PMC3504680
- DOI: 10.1261/rna.035865.112
Alternative transcription start site selection leads to large differences in translation activity in yeast
Abstract
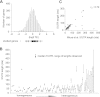
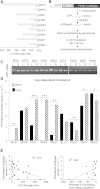
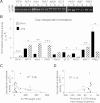
mRNA levels do not accurately predict protein levels in eukaryotic cells. To investigate contributions of 5' untranslated regions (5' UTRs) to mRNA-specific differences in translation, we determined the 5' UTR boundaries of 96 yeast genes for which in vivo translational efficiency varied by 80-fold. A total of 25% of genes showed substantial 5' UTR heterogeneity. We compared the capacity of these genes' alternative 5' UTR isoforms for cap-dependent and cap-independent translation using quantitative in vitro and in vivo translation assays. Six out of nine genes showed mRNA isoform-specific translation activity differences of greater than threefold in at least one condition. For three genes, in vivo translation activities of alternative 5' UTR isoforms differed by more than 100-fold. These results show that changing genes' 5' UTR boundaries can produce large changes in protein output without changing the overall amount of mRNA. Because transcription start site (TSS) heterogeneity is common, we suggest that TSS choice is greatly under-appreciated as a quantitatively significant mechanism for regulating protein production.
Figures


Similar articles
-
Poly(A)-tail-promoted translation in yeast: implications for translational control.RNA. 1998 Nov;4(11):1321-31. doi: 10.1017/s1355838298980669. RNA. 1998. PMID: 9814754 Free PMC article.
-
Cap-independent translation is required for starvation-induced differentiation in yeast.Science. 2007 Aug 31;317(5842):1224-7. doi: 10.1126/science.1144467. Science. 2007. PMID: 17761883
-
Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae.Mol Cell Biol. 1994 Nov;14(11):7322-30. doi: 10.1128/mcb.14.11.7322-7330.1994. Mol Cell Biol. 1994. PMID: 7935446 Free PMC article.
-
Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae.Annu Rev Biochem. 2000;69:571-95. doi: 10.1146/annurev.biochem.69.1.571. Annu Rev Biochem. 2000. PMID: 10966469 Review.
-
Cap-Independent Translation: What's in a Name?Trends Biochem Sci. 2018 Nov;43(11):882-895. doi: 10.1016/j.tibs.2018.04.011. Epub 2018 May 19. Trends Biochem Sci. 2018. PMID: 29789219 Review.
Cited by
-
Non-canonical translation initiation in yeast generates a cryptic pool of mitochondrial proteins.Nucleic Acids Res. 2019 Jun 20;47(11):5777-5791. doi: 10.1093/nar/gkz301. Nucleic Acids Res. 2019. PMID: 31216041 Free PMC article.
-
Herbal Extract Mixture Modulates Intestinal Antioxidative Capacity and Microbiota in Weaning Piglets.Front Microbiol. 2021 Jul 28;12:706758. doi: 10.3389/fmicb.2021.706758. eCollection 2021. Front Microbiol. 2021. PMID: 34394056 Free PMC article.
-
Induction of a unique isoform of the NCOA7 oxidation resistance gene by interferon β-1b.J Interferon Cytokine Res. 2015 Mar;35(3):186-99. doi: 10.1089/jir.2014.0115. Epub 2014 Oct 20. J Interferon Cytokine Res. 2015. PMID: 25330068 Free PMC article.
-
Genome-wide characterization of intergenic polyadenylation sites redefines gene spaces in Arabidopsis thaliana.BMC Genomics. 2015 Jul 9;16(1):511. doi: 10.1186/s12864-015-1691-1. BMC Genomics. 2015. PMID: 26155789 Free PMC article.
-
So close, no matter how far: multiple paths connecting transcription to mRNA translation in eukaryotes.EMBO Rep. 2020 Sep 3;21(9):e50799. doi: 10.15252/embr.202050799. Epub 2020 Aug 16. EMBO Rep. 2020. PMID: 32803873 Free PMC article. Review.
References
-
- Berthelot K, Muldoon M, Rajkowitsch L, Hughes J, McCarthy JE 2004. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol Microbiol 51: 987–1001 - PubMed
-
- Beyer A, Hollunder J, Nasheuer HP, Wilhelm T 2004. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol Cell Proteomics 3: 1083–1092 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
