CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters
- PMID: 23100115
- PMCID: PMC3514669
- DOI: 10.1101/gr.138776.112
CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters
Abstract
One clear hallmark of mammalian promoters is the presence of CpG islands (CGIs) at more than two-thirds of genes, whereas TATA boxes are only present at a minority of promoters. Using genome-wide approaches, we show that GC content and CGIs are major promoter elements in mammalian cells, able to govern open chromatin conformation and support paused transcription. First, we define three classes of promoters with distinct transcriptional directionality and pausing properties that correlate with their GC content. We further analyze the direct influence of GC content on nucleosome positioning and depletion and show that CpG content and CGI width correlate with nucleosome depletion both in vivo and in vitro. We also show that transcription is not essential for nucleosome exclusion but influences both a weak +1 and a well-positioned nucleosome at CGI borders. Altogether our data support the idea that CGIs have become an essential feature of promoter structure defining novel regulatory properties in mammals.
Figures
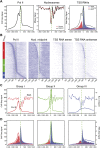
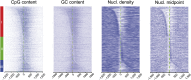
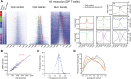
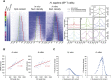

Similar articles
-
Promoter-Targeted Small Activating RNAs Alter Nucleosome Positioning.Adv Exp Med Biol. 2017;983:53-61. doi: 10.1007/978-981-10-4310-9_4. Adv Exp Med Biol. 2017. PMID: 28639191
-
Transcription factor binding site positioning in yeast: proximal promoter motifs characterize TATA-less promoters.PLoS One. 2011;6(9):e24279. doi: 10.1371/journal.pone.0024279. Epub 2011 Sep 9. PLoS One. 2011. PMID: 21931670 Free PMC article.
-
Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae.Mol Biol Cell. 2009 Aug;20(15):3503-13. doi: 10.1091/mbc.e09-02-0111. Epub 2009 Jun 3. Mol Biol Cell. 2009. PMID: 19494041 Free PMC article.
-
CpG islands and the regulation of transcription.Genes Dev. 2011 May 15;25(10):1010-22. doi: 10.1101/gad.2037511. Genes Dev. 2011. PMID: 21576262 Free PMC article. Review.
-
Sequence determinants, function, and evolution of CpG islands.Biochem Soc Trans. 2021 Jun 30;49(3):1109-1119. doi: 10.1042/BST20200695. Biochem Soc Trans. 2021. PMID: 34156435 Free PMC article. Review.
Cited by
-
On the irrelevancy of hydroxyl radical to DNA damage from oxidative stress and implications for epigenetics.Chem Soc Rev. 2020 Sep 21;49(18):6524-6528. doi: 10.1039/d0cs00579g. Chem Soc Rev. 2020. PMID: 32785348 Free PMC article. Review.
-
A widespread role of the motif environment in transcription factor binding across diverse protein families.Genome Res. 2015 Sep;25(9):1268-80. doi: 10.1101/gr.184671.114. Epub 2015 Jul 9. Genome Res. 2015. PMID: 26160164 Free PMC article.
-
Sequence Characteristics Distinguish Transcribed Enhancers from Promoters and Predict Their Breadth of Activity.Genetics. 2019 Apr;211(4):1205-1217. doi: 10.1534/genetics.118.301895. Epub 2019 Jan 29. Genetics. 2019. PMID: 30696717 Free PMC article.
-
High-sensitive nascent transcript sequencing reveals BRD4-specific control of widespread enhancer and target gene transcription.Nat Commun. 2023 Aug 17;14(1):4971. doi: 10.1038/s41467-023-40633-y. Nat Commun. 2023. PMID: 37591883 Free PMC article.
-
Fine-tuning of epigenetic regulation with respect to promoter CpG content in a cell type-specific manner.Epigenetics. 2014 May;9(5):747-59. doi: 10.4161/epi.28075. Epub 2014 Feb 12. Epigenetics. 2014. PMID: 24521667 Free PMC article.
References
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
