Structural insight into HIV-1 capsid recognition by rhesus TRIM5α
- PMID: 23091002
- PMCID: PMC3494900
- DOI: 10.1073/pnas.1210903109
Structural insight into HIV-1 capsid recognition by rhesus TRIM5α
Abstract
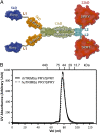
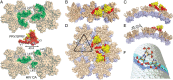
Tripartite motif protein isoform 5 alpha (TRIM5α) is a potent antiviral protein that restricts infection by HIV-1 and other retroviruses. TRIM5α recognizes the lattice of the retrovirus capsid through its B30.2 (PRY/SPRY) domain in a species-specific manner. Upon binding, TRIM5α induces premature disassembly of the viral capsid and activates the downstream innate immune response. We have determined the crystal structure of the rhesus TRIM5α PRY/SPRY domain that reveals essential features for capsid binding. Combined cryo-electron microscopy and biochemical data show that the monomeric rhesus TRIM5α PRY/SPRY, but not the human TRIM5α PRY/SPRY, can bind to HIV-1 capsid protein assemblies without causing disruption of the capsid. This suggests that the PRY/SPRY domain alone constitutes an important pattern-sensing component of TRIM5α that is capable of interacting with viral capsids of different curvatures. Our results provide molecular insights into the mechanisms of TRIM5α-mediated retroviral restriction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.J Virol. 2018 Jan 30;92(4):e01563-17. doi: 10.1128/JVI.01563-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187540 Free PMC article.
-
Rhesus TRIM5α disrupts the HIV-1 capsid at the inter-hexamer interfaces.PLoS Pathog. 2011 Mar;7(3):e1002009. doi: 10.1371/journal.ppat.1002009. Epub 2011 Mar 24. PLoS Pathog. 2011. PMID: 21455494 Free PMC article.
-
Structure of the rhesus monkey TRIM5α PRYSPRY domain, the HIV capsid recognition module.Proc Natl Acad Sci U S A. 2012 Aug 14;109(33):13278-83. doi: 10.1073/pnas.1203536109. Epub 2012 Jul 30. Proc Natl Acad Sci U S A. 2012. PMID: 22847415 Free PMC article.
-
Retroviral restriction factors TRIM5α: therapeutic strategy to inhibit HIV-1 replication.Curr Med Chem. 2011;18(17):2649-54. doi: 10.2174/092986711795933687. Curr Med Chem. 2011. PMID: 21568899 Review.
-
Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence?Immunology. 2005 Dec;116(4):411-7. doi: 10.1111/j.1365-2567.2005.02248.x. Immunology. 2005. PMID: 16313355 Free PMC article. Review.
Cited by
-
A putative SUMO interacting motif in the B30.2/SPRY domain of rhesus macaque TRIM5α important for NF-κB/AP-1 signaling and HIV-1 restriction.Heliyon. 2016 Jan 21;2(1):e00056. doi: 10.1016/j.heliyon.2015.e00056. eCollection 2016 Jan. Heliyon. 2016. PMID: 27441239 Free PMC article.
-
The Three-Fold Axis of the HIV-1 Capsid Lattice Is the Species-Specific Binding Interface for TRIM5α.J Virol. 2018 Feb 12;92(5):e01541-17. doi: 10.1128/JVI.01541-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237846 Free PMC article.
-
Host and viral determinants of Mx2 antiretroviral activity.J Virol. 2014 Jul;88(14):7738-52. doi: 10.1128/JVI.00214-14. Epub 2014 Apr 23. J Virol. 2014. PMID: 24760893 Free PMC article.
-
HIV suppression by host restriction factors and viral immune evasion.Curr Opin Struct Biol. 2015 Apr;31:106-14. doi: 10.1016/j.sbi.2015.04.004. Epub 2015 May 16. Curr Opin Struct Biol. 2015. PMID: 25939065 Free PMC article. Review.
-
Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site.Nat Commun. 2016 Mar 4;7:10714. doi: 10.1038/ncomms10714. Nat Commun. 2016. PMID: 26940118 Free PMC article.
References
-
- Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

