IFN-inducible GTPases in host cell defense
- PMID: 23084913
- PMCID: PMC3490204
- DOI: 10.1016/j.chom.2012.09.007
IFN-inducible GTPases in host cell defense
Abstract
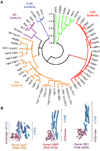
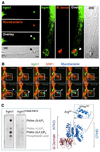
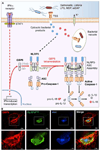
From plants to humans, the ability to control infection at the level of an individual cell-a process termed cell-autonomous immunity-equates firmly with survival of the species. Recent work has begun to unravel this programmed cell-intrinsic response and the central roles played by IFN-inducible GTPases in defending the mammalian cell's interior against a diverse group of invading pathogens. These immune GTPases regulate vesicular traffic and protein complex assembly to stimulate oxidative, autophagic, membranolytic, and inflammasome-related antimicrobial activities within the cytosol, as well as on pathogen-containing vacuoles. Moreover, human genome-wide association studies and disease-related transcriptional profiling have linked mutations in the Immunity-Related GTPase M (IRGM) locus and altered expression of guanylate binding proteins (GBPs) with tuberculosis susceptibility and Crohn's colitis.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
IFN-inducible GTPases and immunity to intracellular pathogens.Trends Immunol. 2004 Nov;25(11):601-9. doi: 10.1016/j.it.2004.08.010. Trends Immunol. 2004. PMID: 15489189 Review.
-
Interferon-Inducible GTPases in Host Resistance, Inflammation and Disease.J Mol Biol. 2016 Aug 28;428(17):3495-513. doi: 10.1016/j.jmb.2016.04.032. Epub 2016 May 12. J Mol Biol. 2016. PMID: 27181197 Free PMC article. Review.
-
Regulation of innate immune functions by guanylate-binding proteins.Int J Med Microbiol. 2018 Jan;308(1):237-245. doi: 10.1016/j.ijmm.2017.10.013. Epub 2017 Nov 2. Int J Med Microbiol. 2018. PMID: 29174633 Review.
-
Interferon-induced GTPases orchestrate host cell-autonomous defence against bacterial pathogens.Biochem Soc Trans. 2021 Jun 30;49(3):1287-1297. doi: 10.1042/BST20200900. Biochem Soc Trans. 2021. PMID: 34003245 Free PMC article. Review.
-
Guanylate Binding Proteins Restrict Leishmania donovani Growth in Nonphagocytic Cells Independent of Parasitophorous Vacuolar Targeting.mBio. 2020 Jul 28;11(4):e01464-20. doi: 10.1128/mBio.01464-20. mBio. 2020. PMID: 32723921 Free PMC article.
Cited by
-
Cell-autonomous Toxoplasma killing program requires Irgm2 but not its microbe vacuolar localization.Life Sci Alliance. 2021 Jun 2;4(7):e202000960. doi: 10.26508/lsa.202000960. Print 2021 Jul. Life Sci Alliance. 2021. PMID: 34078740 Free PMC article.
-
RabGDIα is a negative regulator of interferon-γ-inducible GTPase-dependent cell-autonomous immunity to Toxoplasma gondii.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):E4581-90. doi: 10.1073/pnas.1510031112. Epub 2015 Aug 3. Proc Natl Acad Sci U S A. 2015. PMID: 26240314 Free PMC article.
-
Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses.Retrovirology. 2015 May 16;12:41. doi: 10.1186/s12977-015-0165-5. Retrovirology. 2015. PMID: 25980612 Free PMC article.
-
Initial phospholipid-dependent Irgb6 targeting to Toxoplasma gondii vacuoles mediates host defense.Life Sci Alliance. 2019 Dec 18;3(1):e201900549. doi: 10.26508/lsa.201900549. Print 2020 Jan. Life Sci Alliance. 2019. PMID: 31852733 Free PMC article.
-
Regulation of hippocampal neuronal apoptosis and autophagy in mice with sepsis-associated encephalopathy by immunity-related GTPase M1.CNS Neurosci Ther. 2020 Feb;26(2):177-188. doi: 10.1111/cns.13229. Epub 2019 Oct 14. CNS Neurosci Ther. 2020. PMID: 31612615 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

