Design of peptide-membrane interactions to modulate single-file water transport through modified gramicidin channels
- PMID: 23083713
- PMCID: PMC3475385
- DOI: 10.1016/j.bpj.2012.08.059
Design of peptide-membrane interactions to modulate single-file water transport through modified gramicidin channels
Abstract
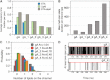
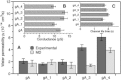
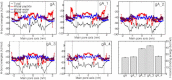
Water permeability through single-file channels is affected by intrinsic factors such as their size and polarity and by external determinants like their lipid environment in the membrane. Previous computational studies revealed that the obstruction of the channel by lipid headgroups can be long-lived, in the range of nanoseconds, and that pore-length-matching membrane mimetics could speed up water permeability. To test the hypothesis of lipid-channel interactions modulating channel permeability, we designed different gramicidin A derivatives with attached acyl chains. By combining extensive molecular-dynamics simulations and single-channel water permeation measurements, we show that by tuning lipid-channel interactions, these modifications reduce the presence of lipid headgroups in the pore, which leads to a clear and selective increase in their water permeability.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Influence of hydrophobic mismatch on structures and dynamics of gramicidin a and lipid bilayers.Biophys J. 2012 Apr 4;102(7):1551-60. doi: 10.1016/j.bpj.2012.03.014. Epub 2012 Apr 3. Biophys J. 2012. PMID: 22500755 Free PMC article.
-
Simulation study of a gramicidin/lipid bilayer system in excess water and lipid. II. Rates and mechanisms of water transport.Biophys J. 1999 Apr;76(4):1939-50. doi: 10.1016/S0006-3495(99)77353-4. Biophys J. 1999. PMID: 10096892 Free PMC article.
-
Invariance of single-file water mobility in gramicidin-like peptidic pores as function of pore length.Biophys J. 2007 Jun 1;92(11):3930-7. doi: 10.1529/biophysj.106.102921. Epub 2007 Mar 16. Biophys J. 2007. PMID: 17369423 Free PMC article.
-
Gramicidin channels.IEEE Trans Nanobioscience. 2005 Mar;4(1):10-20. doi: 10.1109/tnb.2004.842470. IEEE Trans Nanobioscience. 2005. PMID: 15816168 Review.
-
The gramicidin ion channel: a model membrane protein.Biochim Biophys Acta. 2007 Sep;1768(9):2011-25. doi: 10.1016/j.bbamem.2007.05.011. Epub 2007 May 18. Biochim Biophys Acta. 2007. PMID: 17572379 Review.
Cited by
-
Computational analysis of local membrane properties.J Comput Aided Mol Des. 2013 Oct;27(10):845-58. doi: 10.1007/s10822-013-9684-0. Epub 2013 Oct 23. J Comput Aided Mol Des. 2013. PMID: 24150904 Free PMC article.
-
Amino acid motifs in natural products: synthesis of O-acylated derivatives of (2S,3S)-3-hydroxyleucine.Beilstein J Org Chem. 2014 May 16;10:1135-42. doi: 10.3762/bjoc.10.113. eCollection 2014. Beilstein J Org Chem. 2014. PMID: 24991264 Free PMC article.
-
Water Determines the Structure and Dynamics of Proteins.Chem Rev. 2016 Jul 13;116(13):7673-97. doi: 10.1021/acs.chemrev.5b00664. Epub 2016 May 17. Chem Rev. 2016. PMID: 27186992 Free PMC article. Review.
-
Single-file transport of water through membrane channels.Faraday Discuss. 2018 Sep 28;209(0):9-33. doi: 10.1039/c8fd00122g. Faraday Discuss. 2018. PMID: 30014085 Free PMC article.
References
-
- Lee A.G. How lipids and proteins interact in a membrane: a molecular approach. Mol. Biosyst. 2005;1:203–212. - PubMed
-
- Nyholm T.K.M., Ozdirekcan S., Killian J.A. How protein transmembrane segments sense the lipid environment. Biochemistry. 2007;46:1457–1465. - PubMed
-
- Jensen M.O., Mouritsen O.G. Lipids do influence protein function-the hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta. 2004;1666:205–226. - PubMed
-
- Valiyaveetil F.I., Zhou Y., MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

