Pluripotent stem cells escape from senescence-associated DNA methylation changes
- PMID: 23080539
- PMCID: PMC3561866
- DOI: 10.1101/gr.141945.112
Pluripotent stem cells escape from senescence-associated DNA methylation changes
Abstract
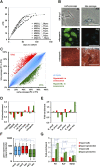
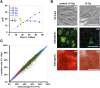
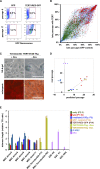
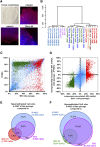
Pluripotent stem cells evade replicative senescence, whereas other primary cells lose their proliferation and differentiation potential after a limited number of cell divisions, and this is accompanied by specific senescence-associated DNA methylation (SA-DNAm) changes. Here, we investigate SA-DNAm changes in mesenchymal stromal cells (MSC) upon long-term culture, irradiation-induced senescence, immortalization, and reprogramming into induced pluripotent stem cells (iPSC) using high-density HumanMethylation450 BeadChips. SA-DNAm changes are highly reproducible and they are enriched in intergenic and nonpromoter regions of developmental genes. Furthermore, SA-hypomethylation in particular appears to be associated with H3K9me3, H3K27me3, and Polycomb-group 2 target genes. We demonstrate that ionizing irradiation, although associated with a senescence phenotype, does not affect SA-DNAm. Furthermore, overexpression of the catalytic subunit of the human telomerase (TERT) or conditional immortalization with a doxycycline-inducible system (TERT and SV40-TAg) result in telomere extension, but do not prevent SA-DNAm. In contrast, we demonstrate that reprogramming into iPSC prevents almost the entire set of SA-DNAm changes. Our results indicate that long-term culture is associated with an epigenetically controlled process that stalls cells in a particular functional state, whereas irradiation-induced senescence and immortalization are not causally related to this process. Absence of SA-DNAm in pluripotent cells may play a central role for their escape from cellular senescence.
Figures
Similar articles
-
Epigenetic rejuvenation of mesenchymal stromal cells derived from induced pluripotent stem cells.Stem Cell Reports. 2014 Sep 9;3(3):414-22. doi: 10.1016/j.stemcr.2014.07.003. Epub 2014 Aug 14. Stem Cell Reports. 2014. PMID: 25241740 Free PMC article.
-
Proof of principle: quality control of therapeutic cell preparations using senescence-associated DNA-methylation changes.BMC Res Notes. 2014 Apr 23;7:254. doi: 10.1186/1756-0500-7-254. BMC Res Notes. 2014. PMID: 24755407 Free PMC article.
-
Senescence-Associated Metabolomic Phenotype in Primary and iPSC-Derived Mesenchymal Stromal Cells.Stem Cell Reports. 2020 Feb 11;14(2):201-209. doi: 10.1016/j.stemcr.2019.12.012. Epub 2020 Jan 23. Stem Cell Reports. 2020. PMID: 31983656 Free PMC article.
-
Human embryonic stem cells: mechanisms to escape replicative senescence?Stem Cell Rev. 2007 Dec;3(4):270-9. doi: 10.1007/s12015-007-9005-x. Stem Cell Rev. 2007. PMID: 18026912 Review.
-
The other side of the coin: mesenchymal stromal cell immortalization beyond evasion of senescence.Hum Cell. 2023 Sep;36(5):1593-1603. doi: 10.1007/s13577-023-00925-3. Epub 2023 Jun 21. Hum Cell. 2023. PMID: 37341871 Review.
Cited by
-
The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation.Nucleic Acids Res. 2016 Dec 15;44(22):10631-10643. doi: 10.1093/nar/gkw802. Epub 2016 Sep 14. Nucleic Acids Res. 2016. PMID: 27634931 Free PMC article.
-
Dietary restriction in senolysis and prevention and treatment of disease.Crit Rev Food Sci Nutr. 2024;64(16):5242-5268. doi: 10.1080/10408398.2022.2153355. Epub 2022 Dec 9. Crit Rev Food Sci Nutr. 2024. PMID: 36484738 Free PMC article. Review.
-
Influence of maternal obesity, diet and exercise on epigenetic regulation of adipocytes.Mol Aspects Med. 2017 Apr;54:37-49. doi: 10.1016/j.mam.2016.10.003. Epub 2016 Nov 4. Mol Aspects Med. 2017. PMID: 27825817 Free PMC article. Review. No abstract available.
-
Why the impact of mechanical stimuli on stem cells remains a challenge.Cell Mol Life Sci. 2018 Sep;75(18):3297-3312. doi: 10.1007/s00018-018-2830-z. Epub 2018 May 4. Cell Mol Life Sci. 2018. PMID: 29728714 Free PMC article. Review.
-
Epigenetic clock analyses of cellular senescence and ageing.Oncotarget. 2016 Feb 23;7(8):8524-31. doi: 10.18632/oncotarget.7383. Oncotarget. 2016. PMID: 26885756 Free PMC article.
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
