Cell type-specific translational profiling in the Xenopus laevis retina
- PMID: 23074098
- PMCID: PMC4536902
- DOI: 10.1002/dvdy.23880
Cell type-specific translational profiling in the Xenopus laevis retina
Abstract
Background: Translating Ribosome Affinity Purification (TRAP), a method recently developed to generate cell type-specific translational profiles, relies on creating transgenic lines of animals in which a tagged ribosomal protein is placed under regulatory control of a cell type-specific promoter. An antibody is then used to affinity purify the tagged ribosomes so that cell type-specific mRNAs can be isolated from whole tissue lysates.
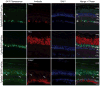
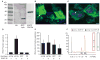
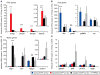
Results: Here, cell type-specific transgenic lines were generated to enable TRAP studies for retinal ganglion cells and rod photoreceptors in the Xenopus laevis retina. Using real time quantitative PCR for assessing expression levels of cell type-specific mRNAs, the TRAP method was shown to selectively isolate mRNAs expressed in the targeted cell and was efficient at purifying mRNAs expressed at both high and low levels. Statistical measures used to distinguish cell type-specific RNAs from low level background and non-specific RNAs showed TRAP to be highly effective in Xenopus.
Conclusions: TRAP can be used to purify mRNAs expressed in rod photoreceptors and retinal ganglion cells in X. laevis. The generated transgenic lines will enable numerous studies into the development, disease, and injury of the X. laevis retina.
Copyright © 2012 Wiley Periodicals, Inc.
Figures

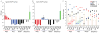

Similar articles
-
Translational profiling of retinal ganglion cell optic nerve regeneration in Xenopus laevis.Dev Biol. 2017 Jun 15;426(2):360-373. doi: 10.1016/j.ydbio.2016.06.003. Epub 2016 Jul 26. Dev Biol. 2017. PMID: 27471010 Free PMC article.
-
FIZ1 is expressed during photoreceptor maturation, and synergizes with NRL and CRX at rod-specific promoters in vitro.Exp Eye Res. 2007 Feb;84(2):349-60. doi: 10.1016/j.exer.2006.10.009. Epub 2006 Dec 4. Exp Eye Res. 2007. PMID: 17141759 Free PMC article.
-
Three cryptochromes are rhythmically expressed in Xenopus laevis retinal photoreceptors.Mol Vis. 2001 Aug 29;7:210-5. Mol Vis. 2001. PMID: 11533577
-
Cytoplasmic mRNA polyadenylation and translation assays.Methods Mol Biol. 2006;322:183-98. doi: 10.1007/978-1-59745-000-3_13. Methods Mol Biol. 2006. PMID: 16739724 Review.
-
Controlling the Messenger: Regulated Translation of Maternal mRNAs in Xenopus laevis Development.Adv Exp Med Biol. 2017;953:49-82. doi: 10.1007/978-3-319-46095-6_2. Adv Exp Med Biol. 2017. PMID: 27975270 Free PMC article. Review.
Cited by
-
Translational profiles of medullary myofibroblasts during kidney fibrosis.J Am Soc Nephrol. 2014 Sep;25(9):1979-90. doi: 10.1681/ASN.2013101143. Epub 2014 Mar 20. J Am Soc Nephrol. 2014. PMID: 24652793 Free PMC article.
-
Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP).Nat Protoc. 2014;9(6):1282-91. doi: 10.1038/nprot.2014.085. Epub 2014 May 8. Nat Protoc. 2014. PMID: 24810037 Free PMC article.
-
Mass spectrometry dataset of LC-MS lipidomics analysis of Xenopus laevis optic nerve.Data Brief. 2023 Jun 14;49:109313. doi: 10.1016/j.dib.2023.109313. eCollection 2023 Aug. Data Brief. 2023. PMID: 37448735 Free PMC article.
-
Dual leucine zipper kinase is necessary for retinal ganglion cell axonal regeneration in Xenopus laevis.PNAS Nexus. 2023 Mar 30;2(5):pgad109. doi: 10.1093/pnasnexus/pgad109. eCollection 2023 May. PNAS Nexus. 2023. PMID: 37152673 Free PMC article.
-
Translational profiling of retinal ganglion cell optic nerve regeneration in Xenopus laevis.Dev Biol. 2017 Jun 15;426(2):360-373. doi: 10.1016/j.ydbio.2016.06.003. Epub 2016 Jul 26. Dev Biol. 2017. PMID: 27471010 Free PMC article.
References
-
- Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21:389–395. - PubMed
-
- Batni S, Scalzetti L, Moody SA, Knox BE. Characterization of the Xenopus rhodopsin gene. J Biol Chem. 1996;271:3179–3186. - PubMed
-
- Calvert PD, Krasnoperova NV, Lyubarsky AL, Isayama T, Nicolo M, Kosaras B, Wong G, Gannon KS, Margolskee RF, Sidman RL, Pugh EN, Jr, Makino CL, Lem J. Phototransduction in transgenic mice after targeted deletion of the rod transducin alpha -subunit. Proc Natl Acad Sci US A. 2000;97:13913–13918. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

