microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.)
- PMID: 23071617
- PMCID: PMC3469577
- DOI: 10.1371/journal.pone.0046703
microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.)
Abstract
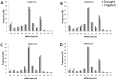
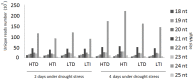
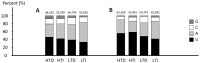
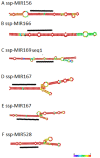
Sugarcane (Saccharum spp.) is one of the most important crops in the world. Drought stress is a major abiotic stress factor that significantly reduces sugarcane yields. However the gene network that mediates plant responses to water stress remains largely unknown in several crop species. Although several microRNAs that mediate post-transcriptional regulation during water stress have been described in other species, the role of the sugarcane microRNAs during drought stress has not been studied. The objective of this work was to identify sugarcane miRNAs that are differentially expressed under drought stress and to correlate this expression with the behavior of two sugarcane cultivars with different drought tolerances. The sugarcane cultivars RB867515 (higher drought tolerance) and RB855536 (lower drought tolerance) were cultivated in a greenhouse for three months and then subjected to drought for 2, 4, 6 or 8 days. By deep sequencing of small RNAs, we were able to identify 18 miRNA families. Among all of the miRNAs thus identified, seven were differentially expressed during drought. Six of these miRNAs were differentially expressed at two days of stress, and five miRNAs were differentially expressed at four days. The expression levels of five miRNAs (ssp-miR164, ssp-miR394, ssp-miR397, ssp-miR399-seq 1 and miR528) were validated by RT-qPCR (quantitative reverse transcriptase PCR). Six precursors and the targets of the differentially expressed miRNA were predicted using an in silico approach and validated by RT-qPCR; many of these targets may play important roles in drought tolerance. These findings constitute a significant increase in the number of identified miRNAs in sugarcane and contribute to the elucidation of the complex regulatory network that is activated by drought stress.
Conflict of interest statement
Figures

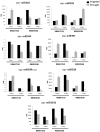
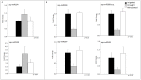
Similar articles
-
Effects of drought on the microtranscriptome of field-grown sugarcane plants.Planta. 2013 Mar;237(3):783-98. doi: 10.1007/s00425-012-1795-7. Epub 2012 Nov 6. Planta. 2013. PMID: 23129215 Free PMC article.
-
Comprehensive transcriptome analysis reveals genes in response to water deficit in the leaves of Saccharum narenga (Nees ex Steud.) hack.BMC Plant Biol. 2018 Oct 20;18(1):250. doi: 10.1186/s12870-018-1428-9. BMC Plant Biol. 2018. PMID: 30342477 Free PMC article.
-
Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane.BMC Plant Biol. 2010 Nov 24;10:260. doi: 10.1186/1471-2229-10-260. BMC Plant Biol. 2010. PMID: 21092324 Free PMC article.
-
Role of microRNAs in plant drought tolerance.Plant Biotechnol J. 2015 Apr;13(3):293-305. doi: 10.1111/pbi.12318. Epub 2015 Jan 13. Plant Biotechnol J. 2015. PMID: 25583362 Free PMC article. Review.
-
MicroRNAs and drought responses in sugarcane.Front Plant Sci. 2015 Feb 23;6:58. doi: 10.3389/fpls.2015.00058. eCollection 2015. Front Plant Sci. 2015. PMID: 25755657 Free PMC article. Review.
Cited by
-
Synthetic versions of firefly luciferase and Renilla luciferase reporter genes that resist transgene silencing in sugarcane.BMC Plant Biol. 2014 Apr 8;14:92. doi: 10.1186/1471-2229-14-92. BMC Plant Biol. 2014. PMID: 24708613 Free PMC article.
-
Drought-Induced miRNA Expression Correlated with Heavy Metal, Phenolic Acid, and Protein and Nitrogen Levels in Five Chickpea Genotypes.ACS Omega. 2023 Sep 18;8(39):35746-35754. doi: 10.1021/acsomega.3c03003. eCollection 2023 Oct 3. ACS Omega. 2023. PMID: 37810661 Free PMC article.
-
Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis.BMC Genomics. 2013 Apr 9;14:233. doi: 10.1186/1471-2164-14-233. BMC Genomics. 2013. PMID: 23570526 Free PMC article.
-
Comparative genome-wide characterization of salt responsive micro RNA and their targets through integrated small RNA and de novo transcriptome profiling in sugarcane and its wild relative Erianthus arundinaceus.3 Biotech. 2024 Jan;14(1):24. doi: 10.1007/s13205-023-03867-7. Epub 2023 Dec 29. 3 Biotech. 2024. PMID: 38162015 Free PMC article.
-
Genome-wide identification of leaf abscission associated microRNAs in sugarcane (Saccharum officinarum L.).BMC Genomics. 2017 Sep 25;18(1):754. doi: 10.1186/s12864-017-4053-3. BMC Genomics. 2017. PMID: 28946845 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

