Selective inhibition of HER2-positive breast cancer cells by the HIV protease inhibitor nelfinavir
- PMID: 23042933
- PMCID: PMC3472971
- DOI: 10.1093/jnci/djs396
Selective inhibition of HER2-positive breast cancer cells by the HIV protease inhibitor nelfinavir
Abstract
Background: Human epidermal growth factor receptor 2 (HER2)-positive breast cancer is highly aggressive and has higher risk of recurrence than HER2-negative cancer. With few treatment options available, new drug targets specific for HER2-positive breast cancer are needed.
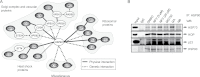
Methods: We conducted a pharmacological profiling of seven genotypically distinct breast cancer cell lines using a subset of inhibitors of breast cancer cells from a screen of the Johns Hopkins Drug Library. To identify molecular targets of nelfinavir, identified in the screen as a selective inhibitor of HER2-positive cells, we conducted a genome-wide screen of a haploinsufficiency yeast mutant collection. We evaluated antitumor activity of nelfinavir with xenografts in athymic nude mouse models (n = 4-6 per group) of human breast cancer and repeated mixed-effects regression analysis. All statistical tests were two-sided.
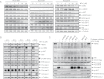
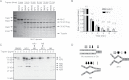
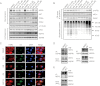
Results: Pharmacological profiling showed that nelfinavir, an anti-HIV drug, selectively inhibited the growth of HER2-positive breast cancer cells in vitro. A genome-wide screening of haploinsufficiency yeast mutants revealed that nelfinavir inhibited heat shock protein 90 (HSP90) function. Further characterization using proteolytic footprinting experiments indicated that nelfinavir inhibited HSP90 in breast cancer cells through a novel mechanism. In vivo, nelfinavir selectively inhibited the growth of HER2-positive breast cancer cells (tumor volume index of HCC1954 cells on day 29, vehicle vs nelfinavir, mean = 14.42 vs 5.16, difference = 9.25, 95% confidence interval [CI] = 5.93 to 12.56, P < .001; tumor volume index of BT474 cells on day 26, vehicle vs nelfinavir, mean = 2.21 vs 0.90, difference = 1.31, 95% CI = 0.83 to 1.78, P < .001). Moreover, nelfinavir inhibited the growth of trastuzumab- and/or lapatinib-resistant, HER2-positive breast cancer cells in vitro at clinically achievable concentrations.
Conclusion: Nelfinavir was found to be a new class of HSP90 inhibitor and can be brought to HER2-breast cancer treatment trials with the same dosage regimen as that used among HIV patients.
Figures


Comment in
-
Re: novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations.J Natl Cancer Inst. 2013 Oct 2;105(19):1512. doi: 10.1093/jnci/djt229. Epub 2013 Sep 6. J Natl Cancer Inst. 2013. PMID: 24013179 No abstract available.
-
Re: selective inhibition of Her2-positive breast cancer cells by the HIV protease inhibitor nelfinavir.J Natl Cancer Inst. 2013 Oct 2;105(19):1515. doi: 10.1093/jnci/djt238. Epub 2013 Sep 19. J Natl Cancer Inst. 2013. PMID: 24052619 No abstract available.
Similar articles
-
Re: selective inhibition of Her2-positive breast cancer cells by the HIV protease inhibitor nelfinavir.J Natl Cancer Inst. 2013 Oct 2;105(19):1515. doi: 10.1093/jnci/djt238. Epub 2013 Sep 19. J Natl Cancer Inst. 2013. PMID: 24052619 No abstract available.
-
Effect of lapatinib on the outgrowth of metastatic breast cancer cells to the brain.J Natl Cancer Inst. 2008 Aug 6;100(15):1092-103. doi: 10.1093/jnci/djn216. Epub 2008 Jul 29. J Natl Cancer Inst. 2008. PMID: 18664652 Free PMC article.
-
Re: novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations.J Natl Cancer Inst. 2013 Oct 2;105(19):1512. doi: 10.1093/jnci/djt229. Epub 2013 Sep 6. J Natl Cancer Inst. 2013. PMID: 24013179 No abstract available.
-
Optimizing outcomes in HER2-positive breast cancer: the molecular rationale.Oncology (Williston Park). 2005 Nov;19(13 Suppl 5):5-16. Oncology (Williston Park). 2005. PMID: 19364051 Review.
-
Lapatinib.Recent Results Cancer Res. 2018;211:19-44. doi: 10.1007/978-3-319-91442-8_2. Recent Results Cancer Res. 2018. PMID: 30069757 Review.
Cited by
-
Identification of anthelmintic parbendazole as a therapeutic molecule for HNSCC through connectivity map-based drug repositioning.Acta Pharm Sin B. 2022 May;12(5):2429-2442. doi: 10.1016/j.apsb.2021.12.005. Epub 2021 Dec 20. Acta Pharm Sin B. 2022. PMID: 35646536 Free PMC article.
-
Synergy between Androgen Receptor Antagonism and Inhibition of mTOR and HER2 in Breast Cancer.Mol Cancer Ther. 2017 Jul;16(7):1389-1400. doi: 10.1158/1535-7163.MCT-17-0111. Epub 2017 May 3. Mol Cancer Ther. 2017. PMID: 28468774 Free PMC article.
-
Drug-Repurposing Screening Identifies a Gallic Acid Binding Site on SARS-CoV-2 Non-structural Protein 7.ACS Pharmacol Transl Sci. 2023 Mar 7;6(4):578-586. doi: 10.1021/acsptsci.2c00225. eCollection 2023 Apr 14. ACS Pharmacol Transl Sci. 2023. PMID: 37082753 Free PMC article.
-
Breast cancer, human immunodeficiency virus and highly active antiretroviral treatment; implications for a high-rate seropositive region.Oncol Rev. 2019 Jan 14;13(1):376. doi: 10.4081/oncol.2019.376. eCollection 2019 Jan 14. Oncol Rev. 2019. PMID: 30713605 Free PMC article.
-
Bruceantin targets HSP90 to overcome resistance to hormone therapy in castration-resistant prostate cancer.Theranostics. 2021 Jan 1;11(2):958-973. doi: 10.7150/thno.51478. eCollection 2021. Theranostics. 2021. PMID: 33391515 Free PMC article.
References
-
- American Cancer Society. Breast cancer facts & figures 2011–2012. American Cancer Society Web site. http://www.cancer.org/Research/CancerFactsFigures/BreastCancerFactsFigur... Accessed September 11, 2012.
-
- Offit K. BRCA mutation frequency and penetrance: new data, old debate J Natl Cancer Inst 2006. 98(23):1675–1677 - PubMed
-
- Lakhani SR, Van De Vijver MJ, Jacquemier J. et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2 J Clin Oncol. 2002. 20(9):2310–2318 - PubMed
-
- Osborne CK. Steroid hormone receptors in breast cancer management Breast Cancer Res Treat. 1998. 51(3):227–238 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

