Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells
- PMID: 23035235
- PMCID: PMC3503047
- DOI: 10.1128/JVI.01104-12
Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells
Abstract
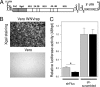
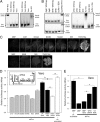
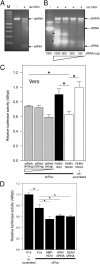
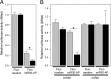
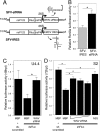
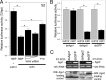
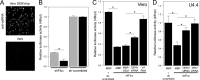
West Nile virus (WNV) and dengue virus (DENV) are highly pathogenic, mosquito-borne flaviviruses (family Flaviviridae) that cause severe disease and death in humans. WNV and DENV actively replicate in mosquitoes and human hosts and thus encounter different host immune responses. RNA interference (RNAi) is the predominant antiviral response against invading RNA viruses in insects and plants. As a countermeasure, plant and insect RNA viruses encode RNA silencing suppressor (RSS) proteins to block the generation/activity of small interfering RNA (siRNA). Enhanced flavivirus replication in mosquitoes depleted for RNAi factors suggests an important biological role for RNAi in restricting virus replication, but it has remained unclear whether or not flaviviruses counteract RNAi via expression of an RSS. First, we established that flaviviral RNA replication suppressed siRNA-induced gene silencing in WNV and DENV replicon-expressing cells. Next, we showed that none of the WNV encoded proteins displayed RSS activity in mammalian and insect cells and in plants by using robust RNAi suppressor assays. In contrast, we found that the 3'-untranslated region-derived RNA molecule known as subgenomic flavivirus RNA (sfRNA) efficiently suppressed siRNA- and miRNA-induced RNAi pathways in both mammalian and insect cells. We also showed that WNV sfRNA inhibits in vitro cleavage of double-stranded RNA by Dicer. The results of the present study suggest a novel role for sfRNA, i.e., as a nucleic acid-based regulator of RNAi pathways, a strategy that may be conserved among flaviviruses.
Figures

Similar articles
-
Zika Virus Subgenomic Flavivirus RNA Generation Requires Cooperativity between Duplicated RNA Structures That Are Essential for Productive Infection in Human Cells.J Virol. 2020 Aug 31;94(18):e00343-20. doi: 10.1128/JVI.00343-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32581095 Free PMC article.
-
Noncoding Subgenomic Flavivirus RNA Is Processed by the Mosquito RNA Interference Machinery and Determines West Nile Virus Transmission by Culex pipiens Mosquitoes.J Virol. 2016 Oct 28;90(22):10145-10159. doi: 10.1128/JVI.00930-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581979 Free PMC article.
-
Pan-flavivirus analysis reveals sfRNA-independent, 3' UTR-biased siRNA production from an insect-specific flavivirus.J Virol. 2024 Nov 19;98(11):e0121524. doi: 10.1128/jvi.01215-24. Epub 2024 Oct 15. J Virol. 2024. PMID: 39404457
-
Functional non-coding RNAs derived from the flavivirus 3' untranslated region.Virus Res. 2015 Aug 3;206:53-61. doi: 10.1016/j.virusres.2015.01.026. Epub 2015 Feb 7. Virus Res. 2015. PMID: 25660582 Review.
-
Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses.Viruses. 2014 Jan 27;6(2):404-27. doi: 10.3390/v6020404. Viruses. 2014. PMID: 24473339 Free PMC article. Review.
Cited by
-
Dicer-2 processes diverse viral RNA species.PLoS One. 2013;8(2):e55458. doi: 10.1371/journal.pone.0055458. Epub 2013 Feb 12. PLoS One. 2013. PMID: 23424633 Free PMC article.
-
Zika Fetal Neuropathogenesis: Etiology of a Viral Syndrome.PLoS Negl Trop Dis. 2016 Aug 25;10(8):e0004877. doi: 10.1371/journal.pntd.0004877. eCollection 2016 Aug. PLoS Negl Trop Dis. 2016. PMID: 27560129 Free PMC article. Review.
-
Different tertiary interactions create the same important 3D features in a distinct flavivirus xrRNA.RNA. 2021 Jan;27(1):54-65. doi: 10.1261/rna.077065.120. Epub 2020 Oct 1. RNA. 2021. PMID: 33004436 Free PMC article.
-
Zika Virus Subgenomic Flavivirus RNA Generation Requires Cooperativity between Duplicated RNA Structures That Are Essential for Productive Infection in Human Cells.J Virol. 2020 Aug 31;94(18):e00343-20. doi: 10.1128/JVI.00343-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32581095 Free PMC article.
-
Potential Application of Drosophila melanogaster as a Model Organism in COVID-19-Related Research.Front Pharmacol. 2020 Sep 4;11:588561. doi: 10.3389/fphar.2020.588561. eCollection 2020. Front Pharmacol. 2020. PMID: 33013425 Free PMC article. No abstract available.
References
-
- Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

