Characterization of de novo synthesized GPCRs supported in nanolipoprotein discs
- PMID: 23028674
- PMCID: PMC3460959
- DOI: 10.1371/journal.pone.0044911
Characterization of de novo synthesized GPCRs supported in nanolipoprotein discs
Abstract
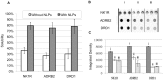
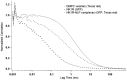
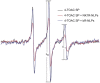
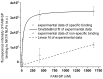
The protein family known as G-protein coupled receptors (GPCRs) comprises an important class of membrane-associated proteins, which remains a difficult family of proteins to characterize because their function requires a native-like lipid membrane environment. This paper focuses on applying a single step method leading to the formation of nanolipoprotein particles (NLPs) capable of solubilizing functional GPCRs for biophysical characterization. NLPs were used to demonstrate increased solubility for multiple GPCRs such as the Neurokinin 1 Receptor (NK1R), the Adrenergic Receptor â2 (ADRB2) and the Dopamine Receptor D1 (DRD1). All three GPCRs showed affinity for their specific ligands using a simple dot blot assay. The NK1R was characterized in greater detail to demonstrate correct folding of the ligand pocket with nanomolar specificity. Electron paramagnetic resonance (EPR) spectroscopy validated the correct folding of the NK1R binding pocket for Substance P (SP). Fluorescence correlation spectroscopy (FCS) was used to identify SP-bound NK1R-containing NLPs and measure their dissociation rate in an aqueous environment. The dissociation constant was found to be 83 nM and was consistent with dot blot assays. This study represents a unique combinational approach involving the single step de novo production of a functional GPCR combined with biophysical techniques to demonstrate receptor association with the NLPs and binding affinity to specific ligands. Such a combined approach provides a novel path forward to screen and characterize GPCRs for drug discovery as well as structural studies outside of the complex cellular environment.
Conflict of interest statement
Figures
Similar articles
-
Single molecule binding of a ligand to a G-protein-coupled receptor in real time using fluorescence correlation spectroscopy, rendered possible by nano-encapsulation in styrene maleic acid lipid particles.Nanoscale. 2020 Jun 4;12(21):11518-11525. doi: 10.1039/d0nr01060j. Nanoscale. 2020. PMID: 32428052
-
Controlling the diameter, monodispersity, and solubility of ApoA1 nanolipoprotein particles using telodendrimer chemistry.Protein Sci. 2013 Aug;22(8):1078-86. doi: 10.1002/pro.2292. Epub 2013 Jun 27. Protein Sci. 2013. PMID: 23754445 Free PMC article.
-
Characterizing diffusion dynamics of a membrane protein associated with nanolipoproteins using fluorescence correlation spectroscopy.Protein Sci. 2011 Feb;20(2):437-47. doi: 10.1002/pro.577. Protein Sci. 2011. PMID: 21280134 Free PMC article.
-
Biophysical characterization of G-protein coupled receptor-peptide ligand binding.Biochem Cell Biol. 2011 Apr;89(2):98-105. doi: 10.1139/o10-142. Biochem Cell Biol. 2011. PMID: 21455262 Free PMC article. Review.
-
Ligands for fluorescence correlation spectroscopy on g protein-coupled receptors.Curr Med Chem. 2012;19(28):4722-30. doi: 10.2174/092986712803341476. Curr Med Chem. 2012. PMID: 22873660 Review.
Cited by
-
Biophysical Characterization of Membrane Proteins Embedded in Nanodiscs Using Fluorescence Correlation Spectroscopy.Membranes (Basel). 2022 Mar 31;12(4):392. doi: 10.3390/membranes12040392. Membranes (Basel). 2022. PMID: 35448362 Free PMC article. Review.
-
Lipid nanoparticle technologies for the study of G protein-coupled receptors in lipid environments.Biophys Rev. 2020 Nov 19;12(6):1287-302. doi: 10.1007/s12551-020-00775-5. Online ahead of print. Biophys Rev. 2020. PMID: 33215301 Free PMC article. Review.
-
Cationic HDL mimetics enhance in vivo delivery of self-replicating mRNA.Nanomedicine. 2020 Feb;24:102154. doi: 10.1016/j.nano.2020.102154. Epub 2020 Jan 24. Nanomedicine. 2020. PMID: 31982617 Free PMC article.
-
Single-Molecule Fluorescence Detection of the Epidermal Growth Factor Receptor in Membrane Discs.Biochemistry. 2019 Jan 29;58(4):286-294. doi: 10.1021/acs.biochem.8b00089. Epub 2018 Apr 6. Biochemistry. 2019. PMID: 29553754 Free PMC article.
-
Cell-Free Co-Translational Approaches for Producing Mammalian Receptors: Expanding the Cell-Free Expression Toolbox Using Nanolipoproteins.Front Pharmacol. 2019 Jul 3;10:744. doi: 10.3389/fphar.2019.00744. eCollection 2019. Front Pharmacol. 2019. PMID: 31333463 Free PMC article. Review.
References
-
- Kostenis E (2004) A glance at G-protein-coupled receptors for lipid mediators: a growing receptor family with remarkably diverse ligands. Pharmacol Ther 102(3): 243–257. - PubMed
-
- Neubig RR, Siderovski DR (2002) Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discovery 1(3): 187–197. - PubMed
-
- Penela P, Ribas C, Mayor F (2003) Mechanisms of regulation of the expression and function of G protein-coupled receptor kinases. Cell Signalling 15(11): 973–981. - PubMed
-
- Lundstrom K (2006) Structural genomics: the ultimate approach for rational drug design. Mol Biotechnol 34(2): 205–212. - PubMed
-
- Lundstrom K (2006) Latest development in drug discovery on G protein-coupled receptors. Curr Protein Pept Sci 7(5): 465–470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

