Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function
- PMID: 23023261
- PMCID: PMC3480716
- DOI: 10.1038/nchembio.1081
Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function
Abstract
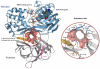
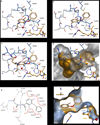
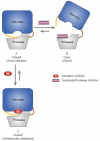
Here we report a highly conserved new binding site located at the interface between the protease and helicase domains of the hepatitis C virus (HCV) NS3 protein. Using a chemical lead, identified by fragment screening and structure-guided design, we demonstrate that this site has a regulatory function on the protease activity via an allosteric mechanism. We propose that compounds binding at this allosteric site inhibit the function of the NS3 protein by stabilizing an inactive conformation and thus represent a new class of direct-acting antiviral agents.
Figures
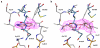
Similar articles
-
Computational study on the inhibitor binding mode and allosteric regulation mechanism in hepatitis C virus NS3/4A protein.PLoS One. 2014 Feb 25;9(2):e87077. doi: 10.1371/journal.pone.0087077. eCollection 2014. PLoS One. 2014. PMID: 24586263 Free PMC article.
-
Allosteric inhibitors of the NS3 protease from the hepatitis C virus.PLoS One. 2013 Jul 30;8(7):e69773. doi: 10.1371/journal.pone.0069773. Print 2013. PLoS One. 2013. PMID: 23936097 Free PMC article.
-
Discovery of Novel Thiophene-Based, Thumb Pocket 2 Allosteric Inhibitors of the Hepatitis C NS5B Polymerase with Improved Potency and Physicochemical Profiles.J Med Chem. 2016 Jul 14;59(13):6293-302. doi: 10.1021/acs.jmedchem.6b00541. Epub 2016 Jul 1. J Med Chem. 2016. PMID: 27366941
-
Hepatitis C virus NS3/4A protease.Antiviral Res. 1999 Feb;41(1):67-84. Antiviral Res. 1999. PMID: 10321580 Review.
-
Novel approaches to the treatment of hepatitis C virus infection.Antivir Chem Chemother. 2000 Mar;11(2):79-96. doi: 10.1177/095632020001100201. Antivir Chem Chemother. 2000. PMID: 10819433 Review.
Cited by
-
Emerging Computational Methods for the Rational Discovery of Allosteric Drugs.Chem Rev. 2016 Jun 8;116(11):6370-90. doi: 10.1021/acs.chemrev.5b00631. Epub 2016 Apr 13. Chem Rev. 2016. PMID: 27074285 Free PMC article. Review.
-
Biophysics in drug discovery: impact, challenges and opportunities.Nat Rev Drug Discov. 2016 Oct;15(10):679-98. doi: 10.1038/nrd.2016.123. Epub 2016 Aug 12. Nat Rev Drug Discov. 2016. PMID: 27516170 Review.
-
Substrate deconstruction and the nonadditivity of enzyme recognition.J Am Chem Soc. 2014 May 21;136(20):7374-82. doi: 10.1021/ja501354q. Epub 2014 May 12. J Am Chem Soc. 2014. PMID: 24791931 Free PMC article.
-
Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19.J Gen Virol. 2021 Mar;102(3):001558. doi: 10.1099/jgv.0.001558. Epub 2021 Jan 28. J Gen Virol. 2021. PMID: 33507143 Free PMC article. Review.
-
Co-evolution networks of HIV/HCV are modular with direct association to structure and function.PLoS Comput Biol. 2018 Sep 7;14(9):e1006409. doi: 10.1371/journal.pcbi.1006409. eCollection 2018 Sep. PLoS Comput Biol. 2018. PMID: 30192744 Free PMC article.
References
-
- Reed KE, Rice CM. Overview of hepatitis c virus genome structure, polyprotein processing and protein properties. Curr. Top. Microbiol. Immunol. 2000;242:55–84. - PubMed
-
- Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J. Gen. Virol. 2000;81:1631–1648. - PubMed
-
- Beran RK, Serebrov V, Pyle AM. The Serine Protease domain of Hepatitis C Viral NS3 Activates RNA Helicase Activity by Promoting the Binding of RNA Substrate. J. Biol. Chem. 2007;282:34913–34920. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- PubChem-Substance/144100018
- PubChem-Substance/144100019
- PubChem-Substance/144100020
- PubChem-Substance/144100021
- PubChem-Substance/144100022
- PubChem-Substance/144100023
- PubChem-Substance/144100024
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

