The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers
- PMID: 23001146
- PMCID: PMC4132664
- DOI: 10.1038/ni.2418
The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers
Abstract
Germinal centers (GCs) are sites of intense B cell proliferation and are central for T cell-dependent antibody responses. However, the role of c-Myc, a key cell-cycle regulator, in this process has been questioned. Here we identified c-Myc(+) B cell subpopulations in immature and mature GCs and found, by genetic ablation of Myc, that they had indispensable roles in the formation and maintenance of GCs. The identification of these functionally critical cellular subsets has implications for human B cell lymphomagenesis, which originates mostly from GC B cells and frequently involves MYC chromosomal translocations. As these translocations are generally dependent on transcription of the recombining partner loci, the c-Myc(+) GC subpopulations may be at a particularly high risk for malignant transformation.
Figures


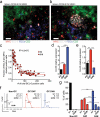
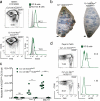

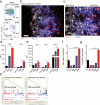
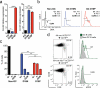
Comment in
-
Tumour immunology: MYCing around with the GC response.Nat Rev Immunol. 2012 Nov;12(11):741. doi: 10.1038/nri3327. Epub 2012 Oct 12. Nat Rev Immunol. 2012. PMID: 23059431 No abstract available.
-
The case of the missing c-Myc.Nat Immunol. 2012 Nov;13(11):1029-31. doi: 10.1038/ni.2455. Nat Immunol. 2012. PMID: 23080195 No abstract available.
Similar articles
-
The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry.Nat Immunol. 2012 Nov;13(11):1083-91. doi: 10.1038/ni.2428. Epub 2012 Sep 23. Nat Immunol. 2012. PMID: 23001145 Free PMC article.
-
The case of the missing c-Myc.Nat Immunol. 2012 Nov;13(11):1029-31. doi: 10.1038/ni.2455. Nat Immunol. 2012. PMID: 23080195 No abstract available.
-
Smc3 dosage regulates B cell transit through germinal centers and restricts their malignant transformation.Nat Immunol. 2021 Feb;22(2):240-253. doi: 10.1038/s41590-020-00827-8. Epub 2021 Jan 11. Nat Immunol. 2021. PMID: 33432228 Free PMC article.
-
New insights into the regulation of human B-cell differentiation.Trends Immunol. 2009 Jun;30(6):277-85. doi: 10.1016/j.it.2009.03.008. Epub 2009 May 15. Trends Immunol. 2009. PMID: 19447676 Free PMC article. Review.
-
MYC's Fine Line Between B Cell Development and Malignancy.Cells. 2020 Feb 24;9(2):523. doi: 10.3390/cells9020523. Cells. 2020. PMID: 32102485 Free PMC article. Review.
Cited by
-
The Transcription Factor AP4 Mediates Resolution of Chronic Viral Infection through Amplification of Germinal Center B Cell Responses.Immunity. 2016 Sep 20;45(3):570-582. doi: 10.1016/j.immuni.2016.07.023. Epub 2016 Aug 23. Immunity. 2016. PMID: 27566940 Free PMC article.
-
rs10924104 in the expression enhancer motif of CD58 confers susceptibility to human autoimmune diseases.Hum Genet. 2024 Jan;143(1):19-33. doi: 10.1007/s00439-023-02617-2. Epub 2023 Nov 23. Hum Genet. 2024. PMID: 37994973
-
Competition for refueling rather than cyclic reentry initiation evident in germinal centers.Sci Immunol. 2022 Mar 11;7(69):eabm0775. doi: 10.1126/sciimmunol.abm0775. Epub 2022 Mar 11. Sci Immunol. 2022. PMID: 35275753 Free PMC article.
-
Cross-talk between Myc and p53 in B-cell lymphomas.Chronic Dis Transl Med. 2019 Oct 22;5(3):139-154. doi: 10.1016/j.cdtm.2019.08.001. eCollection 2019 Sep. Chronic Dis Transl Med. 2019. PMID: 31891126 Free PMC article.
-
Committed Human CD23-Negative Light-Zone Germinal Center B Cells Delineate Transcriptional Program Supporting Plasma Cell Differentiation.Front Immunol. 2021 Dec 2;12:744573. doi: 10.3389/fimmu.2021.744573. eCollection 2021. Front Immunol. 2021. PMID: 34925321 Free PMC article.
References
-
- MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. - PubMed
-
- Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21:2951–2962. - PubMed
-
- de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. - PubMed
-
- Vallespinos M, et al. B Lymphocyte commitment program is driven by the proto-oncogene c-Myc. J Immunol. 2011;186:6726–6736. - PubMed
-
- Shaffer AL, et al. Signatures of the immune response. Immunity. 2001;15:375–385. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

