Genome-wide measurement of RNA folding energies
- PMID: 22981864
- PMCID: PMC3483374
- DOI: 10.1016/j.molcel.2012.08.008
Genome-wide measurement of RNA folding energies
Abstract
RNA structural transitions are important in the function and regulation of RNAs. Here, we reveal a layer of transcriptome organization in the form of RNA folding energies. By probing yeast RNA structures at different temperatures, we obtained relative melting temperatures (Tm) for RNA structures in over 4000 transcripts. Specific signatures of RNA Tm demarcated the polarity of mRNA open reading frames and highlighted numerous candidate regulatory RNA motifs in 3' untranslated regions. RNA Tm distinguished noncoding versus coding RNAs and identified mRNAs with distinct cellular functions. We identified thousands of putative RNA thermometers, and their presence is predictive of the pattern of RNA decay in vivo during heat shock. The exosome complex recognizes unpaired bases during heat shock to degrade these RNAs, coupling intrinsic structural stabilities to gene regulation. Thus, genome-wide structural dynamics of RNA can parse functional elements of the transcriptome and reveal diverse biological insights.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
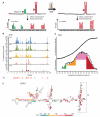
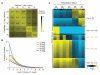
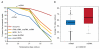
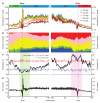
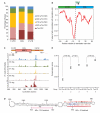
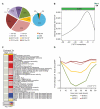
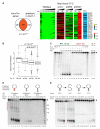
Comment in
-
Gene regulation: RNAs feel the heat.Nat Rev Genet. 2012 Nov;13(11):756. doi: 10.1038/nrg3350. Epub 2012 Sep 25. Nat Rev Genet. 2012. PMID: 23007385 No abstract available.
-
Structure by sequencing: discovery of hidden messages in the noncoding RNA fraction.Mol Cell. 2012 Oct 26;48(2):153-5. doi: 10.1016/j.molcel.2012.10.004. Mol Cell. 2012. PMID: 23102264
Similar articles
-
The mitochondrial RNA landscape of Saccharomyces cerevisiae.PLoS One. 2013 Oct 15;8(10):e78105. doi: 10.1371/journal.pone.0078105. eCollection 2013. PLoS One. 2013. PMID: 24143261 Free PMC article.
-
RNA secondary structure profiling in zebrafish reveals unique regulatory features.BMC Genomics. 2018 Feb 15;19(1):147. doi: 10.1186/s12864-018-4497-0. BMC Genomics. 2018. PMID: 29448945 Free PMC article.
-
CryoEM structure of yeast cytoplasmic exosome complex.Cell Res. 2016 Jul;26(7):822-37. doi: 10.1038/cr.2016.56. Epub 2016 May 13. Cell Res. 2016. PMID: 27174052 Free PMC article.
-
Insights into RNA structure and function from genome-wide studies.Nat Rev Genet. 2014 Jul;15(7):469-79. doi: 10.1038/nrg3681. Epub 2014 May 13. Nat Rev Genet. 2014. PMID: 24821474 Review.
-
The Nuclear RNA Exosome and Its Cofactors.Adv Exp Med Biol. 2019;1203:113-132. doi: 10.1007/978-3-030-31434-7_4. Adv Exp Med Biol. 2019. PMID: 31811632 Review.
Cited by
-
The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly.J Mol Biol. 2013 Oct 23;425(20):3824-38. doi: 10.1016/j.jmb.2013.05.016. Epub 2013 May 28. J Mol Biol. 2013. PMID: 23721653 Free PMC article.
-
SeqFold: genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data.Genome Res. 2013 Feb;23(2):377-87. doi: 10.1101/gr.138545.112. Epub 2012 Oct 11. Genome Res. 2013. PMID: 23064747 Free PMC article.
-
RNA, Action through Interactions.Trends Genet. 2018 Nov;34(11):867-882. doi: 10.1016/j.tig.2018.08.001. Epub 2018 Aug 31. Trends Genet. 2018. PMID: 30177410 Free PMC article. Review.
-
High-throughput determination of RNA structures.Nat Rev Genet. 2018 Oct;19(10):615-634. doi: 10.1038/s41576-018-0034-x. Nat Rev Genet. 2018. PMID: 30054568 Free PMC article. Review.
-
Tackling structures of long noncoding RNAs.Int J Mol Sci. 2013 Dec 4;14(12):23672-84. doi: 10.3390/ijms141223672. Int J Mol Sci. 2013. PMID: 24304541 Free PMC article. Review.
References
-
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell. 2004;15:5–15. - PubMed
-
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

